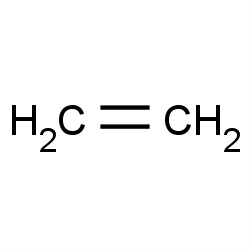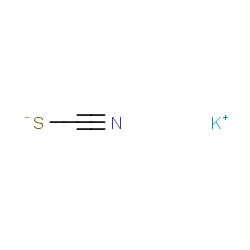complex is
General Information
| Text mining term |
complex is |
| Synonym |
39kD; CI 39kD; Complex I; Complex I 39kD; NADH dehydrogenase (ubiquinone) Fe S protein 2 like; NADH ubiquinone oxidoreductase 39 kDa subunit mitochondrial; NADH ubiquinone oxidoreductase 39 kDa subunit; NDUFA 9… |
| Relative Score |
2127(12,39,53,287) |
| Total Confidence |
1.55 |
No Sequence has found!
Related Compounds
| Compound Name |
Structure |
RScore(About this table) |
|
| paraquat |

|
492(3,10,12,32) |
Details |
| pentachlorophenol |

|
163(2,2,2,3) |
Details |
| atrazine |

|
162(2,2,2,2) |
Details |
| phosphine |

|
238(1,5,6,33) |
Details |
| lindane |
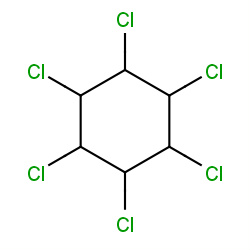
|
112(1,2,2,2) |
Details |
| azobenzene |
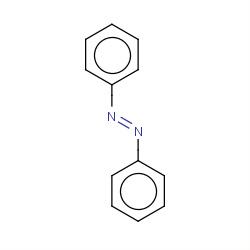
|
95(1,1,1,15) |
Details |
| dieldrin |
|
82(1,1,1,2) |
Details |
| MAA |

|
82(1,1,1,2) |
Details |
| anthraquinone |
|
105(0,3,3,15) |
Details |
| strychnine |
|
98(0,3,3,8) |
Details |
| naphthalene |
|
86(0,2,4,16) |
Details |
| ethylene |
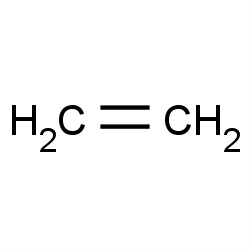
|
36(0,1,1,6) |
Details |
| silica gel |
No Structure Image
|
34(0,1,1,4) |
Details |
| 1-naphthol |

|
32(0,1,1,2) |
Details |
| acrylonitrile |
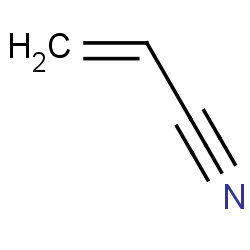
|
32(0,1,1,2) |
Details |
| endosulfan |
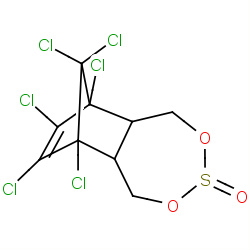
|
31(0,1,1,1) |
Details |
| chloroform |
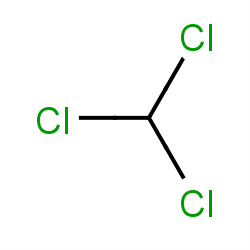
|
26(0,0,2,16) |
Details |
| OCH |

|
15(0,0,1,10) |
Details |
| copper sulfate |

|
11(0,0,0,11) |
Details |
| sulfoxide |
|
10(0,0,0,10) |
Details |
| thiabendazole |
|
10(0,0,1,5) |
Details |
| diphenylamine |
|
8(0,0,0,8) |
Details |
| potassium thiocyanate |
|
8(0,0,1,3) |
Details |
| parathion |
|
7(0,0,1,2) |
Details |
| metiram |

|
6(0,0,1,1) |
Details |
| cresol |

|
5(0,0,0,5) |
Details |
| cyanamide |

|
2(0,0,0,2) |
Details |
| chlorpyrifos |
|
2(0,0,0,2) |
Details |
| dehydroacetic acid |
No Structure Image
|
2(0,0,0,2) |
Details |
| xylenols |
|
1(0,0,0,1) |
Details |
| etofenprox |

|
1(0,0,0,1) |
Details |
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;
 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;