P14
General Information
| Text mining term | P14 |
|---|---|
| Synonym | HSPC003; p14; Late endosomal/lysosomal Mp1 interacting protein; MAPBPIP; Mitogen activated protein binding protein interacting protein |
| Relative Score | 56(0,0,6,26) |
| Total Confidence | -0.15 |
Sequence (Source: NCBI Gene,UniProt)
| ID | Name | GI Number | UniProt | Organism |
|---|---|---|---|---|
| 15998 | Fatty acid-binding protein, liver | 6978825 | FABPL_RAT | Rattus norvegicus |
| 16001 | Fatty acid-binding protein, liver | 6978825 | FABPL_RAT | Rattus norvegicus |
| 15995 | Fatty acid-binding protein, liver | 6978825 | FABPL_RAT | Rattus norvegicus |
| 16003 | Protein S100-A9 | 16758364 | S10A9_RAT | Rattus norvegicus |
| 15999 | Activated RNA polymerase II transcriptional coactivator p15 | 6755364 | TCP4_MOUSE | Mus musculus |
| 16002 | Activated RNA polymerase II transcriptional coactivator p15 | 6755364 | TCP4_MOUSE | Mus musculus |
| 15997 | Activated RNA polymerase II transcriptional coactivator p15 | 57526804 | TCP4_RAT | Rattus norvegicus |
| 16000 | Activated RNA polymerase II transcriptional coactivator p15 | 57526804 | TCP4_RAT | Rattus norvegicus |
| 15996 | Activated RNA polymerase II transcriptional coactivator p15 | 6755364 | TCP4_MOUSE | Mus musculus |
| 15994 | Activated RNA polymerase II transcriptional coactivator p15 | 6755364 | TCP4_MOUSE | Mus musculus |
| ID | Name | GI Number | UniProt | Organism |
Related Compounds
| Compound Name | Structure | RScore(About this table) | |
|---|---|---|---|
| allethrin |

|
7(0,0,1,2) | Details |
| phenothrin |

|
7(0,0,1,2) | Details |
| prallethrin |

|
7(0,0,1,2) | Details |
| flucythrinate |
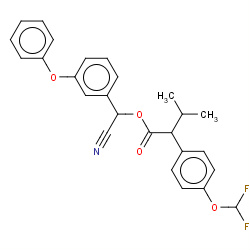
|
7(0,0,1,2) | Details |
| fluvalinate |

|
7(0,0,1,2) | Details |
| strychnine |
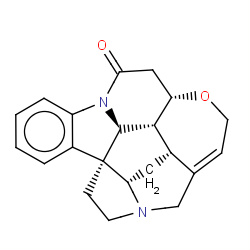
|
4(0,0,0,4) | Details |
| TCA |

|
4(0,0,0,4) | Details |
 Network initializing...Please wait
Network initializing...Please wait
Node List
| ID | Name | Type |
|---|---|---|
Legend:
 : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;