chloride channel (protein family or complex)
General Information
| Text mining term | chloride channel (protein family or complex) |
|---|---|
| Synonym | chloride channel |
| Relative Score | 5998(42,111,125,498) |
| Total Confidence | 4.72 |
Sequence (Source: NCBI Gene,UniProt)
| ID | Name | GI Number | UniProt | Organism |
|---|---|---|---|---|
| 3522 | Glutamate-gated chloride channel subunit | 313849052 | E3WEB4_TETUR | Tetranychus urticae |
| 2719 | GABA-gated chloride channel isoform a3 | 2245681 | O18471_HELVI | Heliothis virescens |
| 2725 | PREDICTED: epithelial chloride channel protein-like | 328707920 | Sternorrhyncha | |
| 3834 | PREDICTED: epithelial chloride channel protein-like | 328707920 | Bellis perennis | |
| 2721 | Gamma-aminobutyric acid receptor subunit beta | 120781 | GBRB_LYMST | Lymnaea stagnalis |
| 3278 | Putative uncharacterized protein | 322796366 | E9IKJ6_SOLIN | Solenopsis invicta |
| 2720 | Histamine-gated chloride channel | 39981112 | Q6TF31_DROSI | Drosophila simulans |
| 3524 | Glutamate-gated chloride channel subunit | 313849052 | E3WEB4_TETUR | Tetranychus urticae |
| 2724 | PREDICTED: epithelial chloride channel protein-like | 328707920 | Sisymbrium irio | |
| 34439 | Chloride channel, putative; Gef1p | 321263915 | E6RDI6_CRYGW | Cryptococcus gattii WM276 |
| 2722 | Glutamate-gated chloride channel | 62901525 | GLUCL_DROME | Drosophila melanogaster |
| 3277 | Glutamate-gated chloride channel subunit | 313849052 | E3WEB4_TETUR | Tetranychus urticae |
| 2723 | Glutamate-gated chloride channel | 247720066 | C6KI52_PLUXY | Plutella xylostella |
| 3523 | Chloride channel protein 7 | 307201523 | E2BSP0_HARSA | Harpegnathos saltator |
| 3279 | Glutamate-gated chloride channel subunit alpha | 109138371 | Q155F9_LOCMI | Locusta migratoria |
| ID | Name | GI Number | UniProt | Organism |
Related Compounds
| Compound Name | Structure | RScore(About this table) | |
|---|---|---|---|
| fipronil |
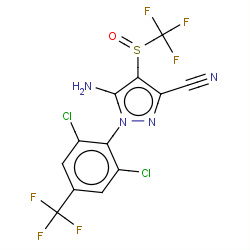
|
762(8,10,13,47) | Details |
| lindane |
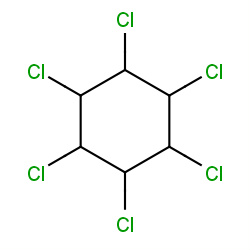
|
709(8,9,9,39) | Details |
| dieldrin |
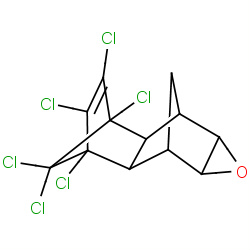
|
617(5,11,12,32) | Details |
| strychnine |
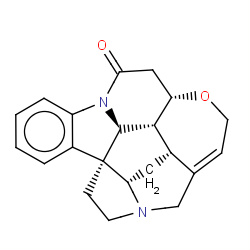
|
530(3,10,13,65) | Details |
| endosulfan |
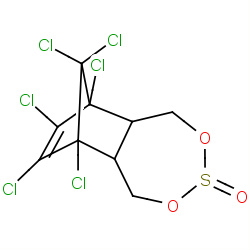
|
179(2,2,2,19) | Details |
| aldrin |
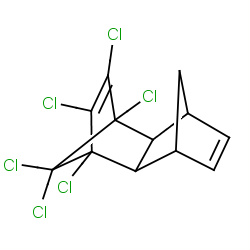
|
163(2,2,2,3) | Details |
| diphenylamine |
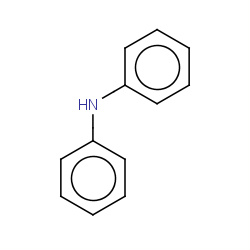
|
1184(1,35,36,79) | Details |
| deltamethrin |

|
156(1,3,4,11) | Details |
| cypermethrin |

|
120(1,2,2,10) | Details |
| fenpropathrin |

|
114(1,2,2,4) | Details |
| tefluthrin |

|
84(1,1,1,4) | Details |
| cyfluthrin |

|
83(1,1,1,3) | Details |
| tetramethrin |
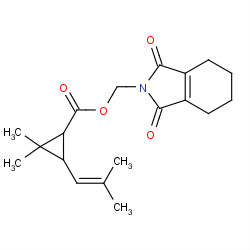
|
33(0,1,1,3) | Details |
| piperazine |
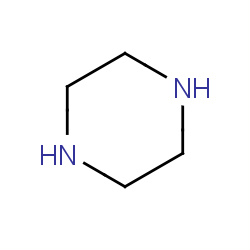
|
32(0,1,1,2) | Details |
| potassium thiocyanate |
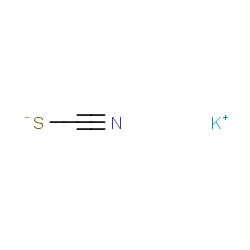
|
32(0,1,1,2) | Details |
| scilliroside |

|
31(0,1,1,1) | Details |
| alpha-cypermethrin |

|
31(0,1,1,1) | Details |
| IAA |
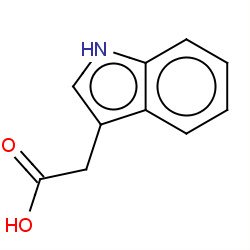
|
36(0,0,0,36) | Details |
| HCH | No Structure Image | 14(0,0,0,14) | Details |
| gamma-HCH | No Structure Image | 14(0,0,0,14) | Details |
| abamectin |
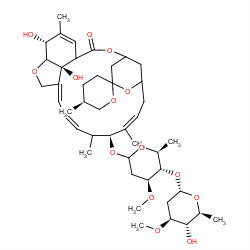
|
10(0,0,0,10) | Details |
| cismethrin |

|
7(0,0,0,7) | Details |
| pyrethrins |
|
6(0,0,0,6) | Details |
| sulfoxide |
|
6(0,0,0,6) | Details |
| piperonyl butoxide |
|
5(0,0,0,5) | Details |
| rotenone |
|
3(0,0,0,3) | Details |
| isobenzan |
|
1(0,0,0,1) | Details |
 Network initializing...Please wait
Network initializing...Please wait
Node List
| ID | Name | Type |
|---|---|---|
Legend:
 : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;