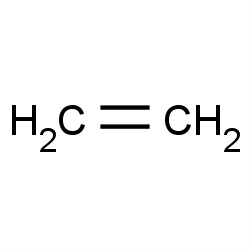Cl 2
General Information
| Text mining term |
Cl 2 |
| Synonym |
AMB11; MHC class I NK cell receptor; CD158E1; CD158E1/2; CD158E2; CL 11; CL 2; HLA BW4 specific inhibitory NK cell receptor… |
| Relative Score |
2542(17,39,71,362) |
| Total Confidence |
1.89 |
No Sequence has found!
Related Compounds
| Compound Name |
Structure |
RScore(About this table) |
|
| phosphine |

|
717(7,10,12,57) |
Details |
| permethrin |

|
253(3,3,3,13) |
Details |
| dithioether |

|
172(2,2,3,7) |
Details |
| dimethenamid |

|
120(1,2,3,5) |
Details |
| carbon disulfide |

|
83(1,1,1,3) |
Details |
| paraquat |

|
82(1,1,1,2) |
Details |
| nitrostyrene |
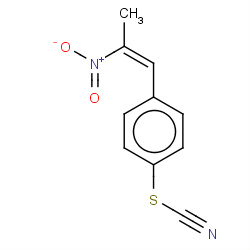
|
82(1,1,1,2) |
Details |
| DDT |
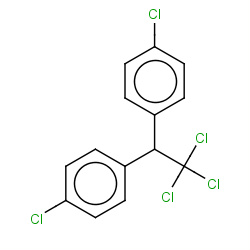
|
81(1,1,1,1) |
Details |
| naphthalene |
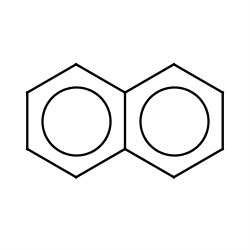
|
143(0,4,5,18) |
Details |
| chloroform |
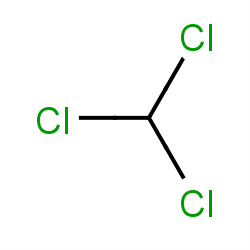
|
81(0,2,3,16) |
Details |
| piperazine |
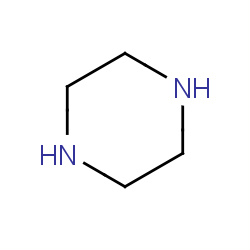
|
75(0,2,3,10) |
Details |
| ethylene oxide |
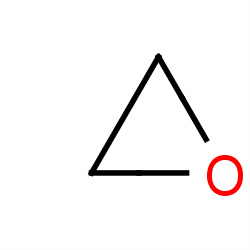
|
63(0,2,2,3) |
Details |
| OCH |

|
82(0,1,3,42) |
Details |
| carbon tetrachloride |
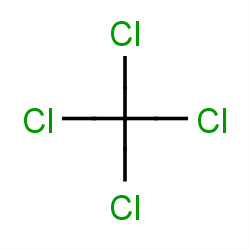
|
46(0,1,2,11) |
Details |
| phosphorus |

|
40(0,1,2,5) |
Details |
| chloranil |
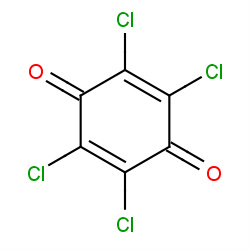
|
38(0,1,2,3) |
Details |
| sulfuric acid |

|
38(0,1,1,8) |
Details |
| cyanamide |

|
35(0,1,1,5) |
Details |
| phthalide |
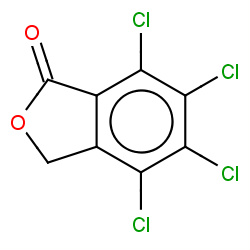
|
31(0,1,1,1) |
Details |
| methylene chloride |
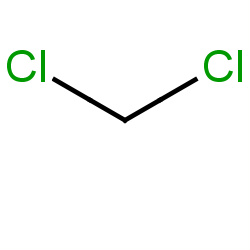
|
103(0,0,9,58) |
Details |
| ethylene |
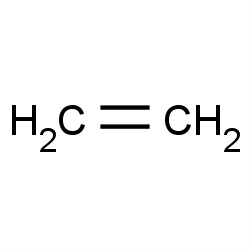
|
30(0,0,4,10) |
Details |
| sulfur |

|
24(0,0,1,19) |
Details |
| butylamine |

|
14(0,0,2,4) |
Details |
| pyrethrins |
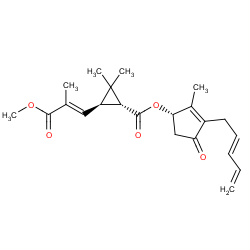
|
13(0,0,0,13) |
Details |
| strychnine |
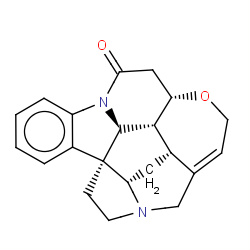
|
12(0,0,1,7) |
Details |
| cresol |

|
8(0,0,1,3) |
Details |
| copper sulfate |

|
7(0,0,1,2) |
Details |
| tau-fluvalinate |

|
6(0,0,0,6) |
Details |
| TCA |

|
6(0,0,0,6) |
Details |
| benzyl benzoate |
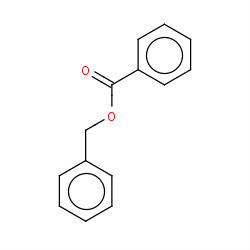
|
6(0,0,1,1) |
Details |
| alpha-cypermethrin |

|
6(0,0,0,6) |
Details |
| sulfoxide |
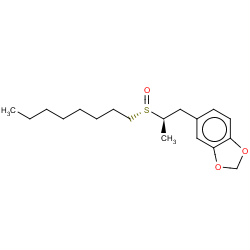
|
6(0,0,0,6) |
Details |
| α-chlorohydrin |
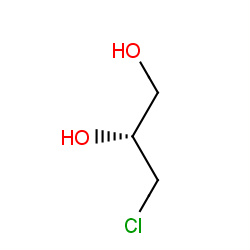
|
3(0,0,0,3) |
Details |
| carpropamid |
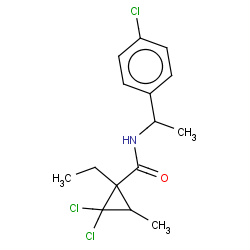
|
1(0,0,0,1) |
Details |
| parathion |
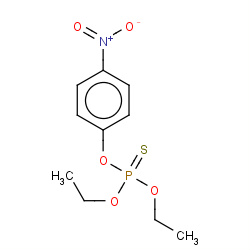
|
1(0,0,0,1) |
Details |
| difenzoquat |

|
1(0,0,0,1) |
Details |
| pyrifenox |
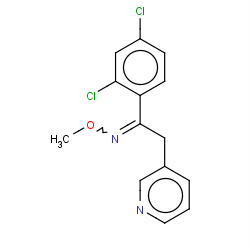
|
1(0,0,0,1) |
Details |
| chlorpyrifos |
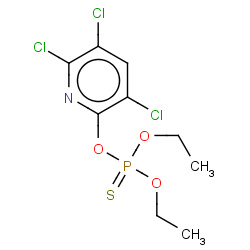
|
1(0,0,0,1) |
Details |
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;
 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;