chloroform
Identification (Source:EPA,EU)
| ID |
1393 |
| Name |
chloroform |
| CAS |
67-66-3 |
| IUPAC |
chloroform |
| Inchi |
InChI=1S/CHCl3/c2-1(3)4/h1H |
| CIPAC |
|
| EPA Code |
|
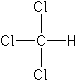
| Formula |
CHCl3 |
| SMILES |
ClC([H])(Cl)Cl |
| Status |
Not Available |
Classification
Properties (Source:PubChem)
| Molecular Weight |
119.37764 |
| Physical State |
|
| AlogP |
1.617 |
| H-Bond Donor |
0 |
| H-Bond Acceptor |
0 |
| Surface Area |
97.61 |
| Polar Surface Area |
0.0 |
Structure
Related targets
| Target Name |
RScore (About this table) |
|
|
Tyrosinase
|
218(2,3,4,23) |
Details |
|
beta adrenoceptors (protein family or complex)
|
174(2,2,4,4) |
Details |
|
TRPC5
|
172(2,2,3,7) |
Details |
|
Glut 4
|
166(2,2,2,6) |
Details |
|
laminin (protein family or complex)
|
163(2,2,2,3) |
Details |
|
Lipoxygenase (protein family or complex)
|
155(1,3,4,10) |
Details |
|
c Myc
|
111(1,1,3,21) |
Details |
|
caspase 3
|
110(1,1,3,20) |
Details |
|
xanthine oxidase
|
94(1,1,1,14) |
Details |
|
Acetylcholinesterase
|
90(1,1,1,10) |
Details |
|
VEGF
|
90(1,1,1,10) |
Details |
|
PdCl 2
|
85(1,1,1,5) |
Details |
|
CCl 3
|
83(1,1,1,3) |
Details |
|
NL4
|
83(1,1,1,3) |
Details |
|
Peroxisome proliferator activated receptors
|
82(1,1,1,2) |
Details |
|
LH2
|
82(1,1,1,2) |
Details |
|
N methyl D aspartate receptors (protein family or complex)
|
82(1,1,1,2) |
Details |
|
alpha 2 adrenergic
|
81(1,1,1,1) |
Details |
|
C21
|
81(1,1,1,1) |
Details |
|
Cl 6
|
81(1,1,1,1) |
Details |
|
whistle
|
81(1,1,1,1) |
Details |
|
MR 1
|
81(1,1,1,1) |
Details |
|
peroxisome proliferator activated receptor gamma
|
131(0,4,4,11) |
Details |
|
butyrylcholinesterase
|
84(0,2,4,14) |
Details |
|
Cl 2
|
81(0,2,3,16) |
Details |
|
interleukin 6
|
72(0,2,2,12) |
Details |
|
interleukin 1 beta
|
71(0,2,2,11) |
Details |
|
PI3K
|
67(0,2,2,7) |
Details |
|
TRIM
|
62(0,2,2,2) |
Details |
|
C18
|
75(0,1,4,30) |
Details |
|
cytochrome P450 2E1
|
49(0,1,2,14) |
Details |
|
lysozyme
|
46(0,1,2,11) |
Details |
|
APC (protein family or complex)
|
40(0,1,1,10) |
Details |
|
tumor necrosis factor alpha
|
39(0,1,2,4) |
Details |
|
ATPase
|
39(0,1,1,9) |
Details |
|
serotonin transporter
|
37(0,1,1,7) |
Details |
|
cytochrome P450 (protein family or complex)
|
36(0,1,1,6) |
Details |
|
cytochrome c
|
36(0,1,1,6) |
Details |
|
CYP3A
|
35(0,1,1,5) |
Details |
|
vascular endothelial growth factor receptor 1
|
35(0,1,1,5) |
Details |
|
protein tyrosine phosphatase 1B
|
35(0,1,1,5) |
Details |
|
PF6
|
34(0,1,1,4) |
Details |
|
apolipoprotein E
|
34(0,1,1,4) |
Details |
|
phosphatidylinositol 3 kinase (protein family or complex)
|
34(0,1,1,4) |
Details |
|
PALS
|
34(0,1,1,4) |
Details |
|
lactate dehydrogenase (protein family or complex)
|
33(0,1,1,3) |
Details |
|
dopamine transporter
|
33(0,1,1,3) |
Details |
|
ICAM 1
|
33(0,1,1,3) |
Details |
|
norepinephrine transporter
|
33(0,1,1,3) |
Details |
|
LPP1
|
33(0,1,1,3) |
Details |
|
interleukin (protein family or complex)
|
33(0,1,1,3) |
Details |
|
BRIC
|
32(0,1,1,2) |
Details |
|
component II
|
32(0,1,1,2) |
Details |
|
glycine receptors (protein family or complex)
|
32(0,1,1,2) |
Details |
|
placental growth factor
|
31(0,1,1,1) |
Details |
|
LH3
|
31(0,1,1,1) |
Details |
|
RP18
|
31(0,1,1,1) |
Details |
|
lipase a
|
31(0,1,1,1) |
Details |
|
A25
|
31(0,1,1,1) |
Details |
|
beta 2 adrenoceptor
|
31(0,1,1,1) |
Details |
|
Dragon
|
31(0,1,1,1) |
Details |
|
MP2
|
30(0,0,2,20) |
Details |
|
complex is
|
26(0,0,2,16) |
Details |
|
Pr 3
|
23(0,0,2,13) |
Details |
|
hemoglobin (protein family or complex)
|
21(0,0,2,11) |
Details |
|
EGFR
|
20(0,0,0,20) |
Details |
|
P glycoprotein
|
16(0,0,1,11) |
Details |
|
c erbB 2
|
16(0,0,0,16) |
Details |
|
alanine aminotransferase
|
14(0,0,1,9) |
Details |
|
PC 3
|
14(0,0,1,9) |
Details |
|
L17
|
13(0,0,1,8) |
Details |
|
BTHS
|
12(0,0,1,7) |
Details |
|
Ca II
|
11(0,0,1,6) |
Details |
|
LAMP
|
11(0,0,1,6) |
Details |
|
SP C
|
11(0,0,1,6) |
Details |
|
PC3
|
11(0,0,1,6) |
Details |
|
neurotensin
|
11(0,0,0,11) |
Details |
|
albumin
|
11(0,0,0,11) |
Details |
|
glutathione S transferase
|
11(0,0,1,6) |
Details |
|
ENs
|
11(0,0,1,6) |
Details |
|
catalase
|
10(0,0,1,5) |
Details |
|
CARS
|
10(0,0,1,5) |
Details |
|
MSX1
|
10(0,0,1,5) |
Details |
|
RL1
|
10(0,0,1,5) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;