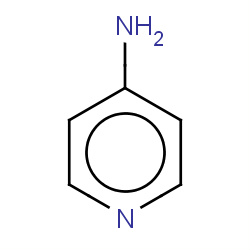
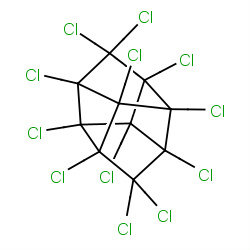
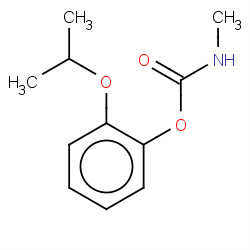
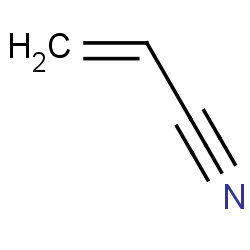
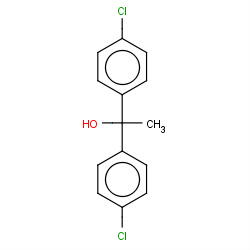
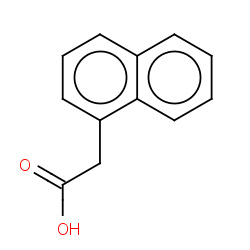
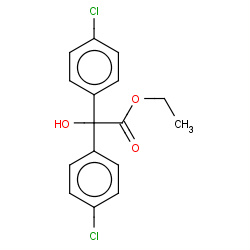
| 22682 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8739 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 72 |
ATP synthase subunit beta |
305671625 |
E0XD18_9APIA |
Hydrocotyle vulgaris
|
| 16586 |
ATPase |
312086086 |
E1G877_LOALO |
Loa loa
|
| 22751 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22815 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8695 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 22903 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 22956 |
ATPase, putative |
58270374 |
Q5KCN0_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 8839 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 22995 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 3874 |
PREDICTED: plasma membrane calcium-transporting ATPase 3-like isoform 1 |
328724399 |
|
Bellis perennis
|
| 8968 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8694 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 3618 |
ATPase beta subunit |
12666551 |
Q9BLQ1_MYZPE |
Myzus persicae
|
| 23061 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 16541 |
ATPase |
312079317 |
E1G060_LOALO |
Loa loa
|
| 23066 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 80 |
ATP synthase subunit beta |
119368040 |
A0ZQC8_OXACO |
Oxalis corniculata
|
| 23059 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8869 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 8966 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 81 |
ATP synthase subunit alpha |
27461567 |
Q8HFD5_TYPLA |
Typha latifolia
|
| 22726 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22753 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 78 |
ATP synthase subunit alpha |
302747606 |
E5DKG6_POROL |
Portulaca oleracea
|
| 22707 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 8895 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22832 |
ATPase, putative |
58271254 |
Q5KC30_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 8757 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 3543 |
Heat shock protein Hsp40 |
91718808 |
Q1PCC3_9MUSC |
Liriomyza huidobrensis
|
| 8921 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 22833 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8777 |
ATPase |
359448103 |
|
Pseudoalteromonas sp. BSi20480
|
| 23065 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8905 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 8984 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 22680 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 22661 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 22961 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 3548 |
V-type proton ATPase subunit B |
401326 |
VATB_HELVI |
Heliothis virescens
|
| 22721 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 8800 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 8791 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 23013 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 2953 |
Sodium/potassium ATPase alpha subunit |
34484237 |
Q6W966_ACRHI |
Acrosternum hilare
|
| 8713 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 8870 |
ATPase |
359449990 |
|
Pseudoalteromonas sp. BSi20480
|
| 8817 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 22976 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 16588 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 22741 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22662 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 22978 |
ATPase, putative |
58270374 |
Q5KCN0_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 69 |
V-type proton ATPase catalytic subunit A |
2493122 |
VATA_BRANA |
Brassica napus
|
| 8714 |
ATPase |
359459786 |
|
Acaryochloris sp. CCMEE 5410
|
| 8754 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 8737 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 8908 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 2938 |
ATPase protein |
285005035 |
D3H5I1_PEA |
Pisum sativum
|
| 22728 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22870 |
ATPase |
331217656 |
E3JYG2_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8752 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 2936 |
Heat shock protein Hsp40 |
91718810 |
Q1PCC2_9MUSC |
Liriomyza sativae
|
| 8802 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 23030 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 8981 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 8691 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 2945 |
ABC membrane transporter |
23379328 |
Q8ISK4_9DIPT |
Tipulidae sp. JK-2001
|
| 22796 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 8989 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 23067 |
ATPase, putative |
58271254 |
Q5KC30_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 23015 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 91 |
ATP synthase subunit beta |
66276285 |
Q29S87_9ROSI |
Marah macrocarpus
|
| 22723 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 22526 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22647 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 22871 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8844 |
ATPase |
359442470 |
|
Pseudoalteromonas sp. BSi20429
|
| 22813 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22994 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 8860 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 22919 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22902 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22681 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8814 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 22939 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22772 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 22630 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 64 |
Calcium ATPase |
289540885 |
D3YBB1_TRIRP |
Trifolium repens
|
| 71 |
ATP synthase subunit c, chloroplastic |
223634981 |
ATPH_LOLPR |
Lolium perenne
|
| 22742 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 23060 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8987 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 8688 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 16563 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 3615 |
Vacuolar ATPase G subunit-like protein |
90820012 |
Q1W2A2_9HEMI |
Graphocephala atropunctata
|
| 2941 |
ATPase subunit 8 |
187370808 |
D7SGR8_9ACAR |
Panonychus ulmi
|
| 22656 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22660 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 88 |
ATP synthase subunit beta |
7706848 |
Q9MRW2_AMAHP |
Amaranthus hypochondriacus
|
| 82 |
ATP synthase subunit beta |
237783999 |
C4PBD1_9SOLA |
Physalis peruviana
|
| 8664 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 3547 |
Heat shock protein Hsp40 |
91718810 |
Q1PCC2_9MUSC |
Liriomyza sativae
|
| 8759 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 2954 |
ATPase ASNA1 homolog |
263406082 |
ASNA_DROSI |
Drosophila simulans
|
| 8841 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 8768 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 8716 |
ATPase |
359456333 |
|
Pseudoalteromonas sp. BSi20495
|
| 22923 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 85 |
ATP synthase beta subunit |
6688490 |
Q9SCB6_LAMAM |
Lamium amplexicaule
|
| 8835 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 22865 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8772 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 22657 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8956 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 2955 |
Plasma membrane calcium ATPase |
119351137 |
A9P669_PINFU |
Pinctada fucata
|
| 22679 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 61 |
ATP synthase subunit alpha |
89112846 |
A1XIS6_9POAL |
Cyperus alternifolius
|
| 3876 |
Sodium/potassium ATPase alpha subunit |
34484237 |
Q6W966_ACRHI |
Acrosternum hilare
|
| 8863 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 2962 |
PREDICTED: plasma membrane calcium-transporting ATPase 3-like isoform 1 |
328724399 |
|
Sternorrhyncha
|
| 22770 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 8770 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22849 |
ATPase |
331217656 |
E3JYG2_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 23029 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8823 |
ATPase |
359459786 |
|
Acaryochloris sp. CCMEE 5410
|
| 8662 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 2958 |
ATPase beta subunit |
12666341 |
Q9BLW7_9HEMI |
Eriosoma lanuginosum
|
| 22866 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22528 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 8667 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8738 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 8820 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 22725 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 62 |
ATP synthase subunit alpha |
334691738 |
F8S0A2_GALAP |
Galium aparine
|
| 8867 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 8939 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8821 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 8707 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 8866 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 22831 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8946 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 8811 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 63 |
ATP synthase subunit alpha |
94317180 |
Q1KZP1_9ROSI |
Erodium pelargoniflorum
|
| 8903 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 22546 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8906 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 23014 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22692 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 8924 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 16754 |
ATPase |
312082008 |
E1G3F4_LOALO |
Loa loa
|
| 8822 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 8840 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 8922 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 22705 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 2942 |
Putative uncharacterized protein |
312285782 |
E5L9V7_BACOL |
Bactrocera oleae
|
| 8838 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 3546 |
ATP-binding cassette sub-family B member 3 |
319894768 |
E9LP53_TRINI |
Trichoplusia ni
|
| 22691 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8755 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 22758 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22867 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 16564 |
ATPase |
312086086 |
E1G877_LOALO |
Loa loa
|
| 59 |
Putative uncharacterized protein |
255032936 |
C7F9Z2_9ASTR |
Lactuca serriola
|
| 77 |
ATPase subunit III |
215982088 |
B8XXA6_9POAL |
Eragrostis pectinacea
|
| 22712 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8673 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 74 |
ATP synthase subunit alpha |
57115599 |
Q5IBI4_9ROSA |
Rubus sp. JPM-2004
|
| 22851 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22754 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22811 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 8880 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 22690 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 8779 |
ATPase |
359456333 |
|
Pseudoalteromonas sp. BSi20495
|
| 8858 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 16753 |
ATPase |
312075807 |
E1FVR9_LOALO |
Loa loa
|
| 22852 |
ATPase, putative |
58266878 |
Q5KI67_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 22696 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 22767 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 16780 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 8661 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 22941 |
ATPase, putative |
58271254 |
Q5KC30_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 8888 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 22769 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22887 |
ATPase, putative |
58264774 |
Q5KKM6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 70 |
ATP-dependent transporter YFL028C |
290574146 |
E9JQ46_SETVI |
Setaria viridis
|
| 8986 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 8847 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 22711 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8798 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 22729 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 54 |
Activator of 90 kDa heat shock protein ATPase |
281399027 |
D2E9R5_DACGL |
Dactylis glomerata
|
| 22628 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22609 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 23058 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22724 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 58 |
Plasma membrane ATPase |
50400847 |
PMA1_WHEAT |
Triticum aestivum
|
| 95 |
ATP synthase subunit beta |
89242548 |
A1XIW2_9LILI |
Najas gracillima
|
| 8979 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8907 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 8965 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 8734 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 22957 |
ATPase |
331217656 |
E3JYG2_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 3549 |
Putative 26S proteasome non-ATPase regulatory subunit 2 |
157813216 |
A9XY44_FORAU |
Forficula auricularia
|
| 3616 |
Putative 26S proteasome non-ATPase regulatory subunit 2 |
157813240 |
A9XY56_CYDPO |
Cydia pomonella
|
| 22922 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 8845 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 8865 |
ATPase |
359442647 |
|
Pseudoalteromonas sp. BSi20429
|
| 22710 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 16750 |
ATPase |
312086086 |
E1G877_LOALO |
Loa loa
|
| 22938 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 22706 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8944 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 22737 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 57 |
ATP synthase subunit c, chloroplastic |
223635051 |
ATPH_CAPBU |
Capsella bursa-pastoris
|
| 22996 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 22587 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8834 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 16751 |
ATPase |
312079317 |
E1G060_LOALO |
Loa loa
|
| 22850 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8894 |
ATPase |
359456333 |
|
Pseudoalteromonas sp. BSi20495
|
| 8670 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 8957 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 22566 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22834 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 22709 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 22812 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8797 |
ATPase |
359442647 |
|
Pseudoalteromonas sp. BSi20429
|
| 60 |
ATP synthase subunit c, chloroplastic |
223635044 |
ATPH_AGRST |
Agrostis stolonifera
|
| 8904 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 16664 |
ATPase |
312086086 |
E1G877_LOALO |
Loa loa
|
| 8889 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 22611 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 22738 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 23051 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8959 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8715 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22508 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8988 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 22586 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8884 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 22629 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 22663 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8693 |
ATPase |
359449990 |
|
Pseudoalteromonas sp. BSi20480
|
| 16589 |
ATPase |
312082008 |
E1G3F4_LOALO |
Loa loa
|
| 22674 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8799 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 22525 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22920 |
ATPase, putative |
58271254 |
Q5KC30_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 56 |
Heat shock protein 83 |
1708314 |
HSP83_IPONI |
Ipomoea nil
|
| 8780 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22524 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22678 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 22584 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 3614 |
Putative vacuolar ATP synthase subunit E |
46561760 |
Q6PPH3_HOMVI |
Homalodisca vitripennis
|
| 22527 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 23033 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 8773 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8753 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 8859 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 16609 |
ATPase |
312082008 |
E1G3F4_LOALO |
Loa loa
|
| 22942 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8885 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 8687 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 16540 |
ATPase |
312074341 |
E1FTW5_LOALO |
Loa loa
|
| 2952 |
Putative uncharacterized protein |
312285782 |
E5L9V7_BACOL |
Bactrocera oleae
|
| 23047 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22665 |
ATPase |
331217656 |
E3JYG2_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22693 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 8891 |
ATPase |
359459786 |
|
Acaryochloris sp. CCMEE 5410
|
| 86 |
Plasma membrane H+-ATPase |
52851188 |
Q5ZFR6_PLAMJ |
Plantago major
|
| 8760 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 8730 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 94 |
ATP synthase subunit beta |
323715506 |
F1JYK6_9ASTE |
Mentzelia dispersa
|
| 8928 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 3545 |
Putative 26S proteasome non-ATPase regulatory subunit 2 |
157813240 |
A9XY56_CYDPO |
Cydia pomonella
|
| 22743 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8682 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 22727 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22568 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 22905 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 8748 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22768 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 22830 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 8886 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 87 |
ATP synthase subunit c, chloroplastic |
223634976 |
ATPH_IPOPU |
Ipomoea purpurea
|
| 8892 |
ATPase |
359448103 |
|
Pseudoalteromonas sp. BSi20480
|
| 23048 |
ATPase, putative |
58266878 |
Q5KI67_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 2948 |
Vacuolar ATPase subunit C |
13173386 |
Q9BMC5_HELAM |
Helicoverpa armigera
|
| 8729 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 8725 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 2944 |
ATP synthase subunit a |
169792304 |
B0L4H3_LOCMI |
Locusta migratoria
|
| 8733 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 16665 |
ATPase |
312077420 |
E1FXT4_LOALO |
Loa loa
|
| 22847 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8776 |
ATPase |
359459786 |
|
Acaryochloris sp. CCMEE 5410
|
| 16666 |
ATPase |
312079317 |
E1G060_LOALO |
Loa loa
|
| 2946 |
Vacuolar ATPase G subunit-like protein |
121543630 |
A2I3V5_MACHI |
Maconellicoccus hirsutus
|
| 8666 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 8967 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 8732 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8794 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 8751 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 22608 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 23062 |
ATPase |
331217656 |
E3JYG2_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8686 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 3877 |
GrpE protein homolog |
11344585 |
Q9GR98_APHGO |
Aphis gossypii
|
| 22708 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 22906 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 2949 |
V-type proton ATPase subunit H |
12585497 |
VATH_MANSE |
Manduca sexta
|
| 8882 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 3551 |
ATPase type 13A1 |
307716047 |
E2JE29_HELZE |
Helicoverpa zea
|
| 76 |
ATP synthase subunit alpha |
6561705 |
Q9T713_ALIPL |
Alisma plantago-aquatica
|
| 8960 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 22642 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 16669 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 22757 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 3878 |
Vacuolar ATPase G subunit-like protein |
90820012 |
Q1W2A2_9HEMI |
Graphocephala atropunctata
|
| 8943 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 8893 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 8769 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 22868 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 22646 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 2939 |
Sodium/potassium ATPase alpha subunit |
34484237 |
Q6W966_ACRHI |
Acrosternum hilare
|
| 8736 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 8704 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 8758 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 8728 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 22869 |
ATPase, putative |
58270374 |
Q5KCN0_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 8923 |
ATPase |
359683273 |
|
Leptospira santarosai str. 2000030832
|
| 79 |
ATP synthase subunit b, chloroplastic |
226741428 |
ATPF_CUSOB |
Cuscuta obtusiflora
|
| 22506 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 22886 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 8711 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 89 |
ATP synthase subunit beta |
14718232 |
Q95FJ7_STEME |
Stellaria media
|
| 2957 |
Vacuolar ATPase G subunit-like protein |
121543630 |
A2I3V5_MACHI |
Maconellicoccus hirsutus
|
| 22759 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8750 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 22664 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8735 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 2934 |
Putative flagellum-specific ATP synthase |
52630923 |
Q5XUC2_TOXCI |
Toxoptera citricida
|
| 22605 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 22771 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8782 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8792 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 83 |
ATPase subunit 6 |
349806554 |
|
Lepidium didymum
|
| 68 |
ATP synthase subunit beta |
110915688 |
A9L9Y3_9POAL |
Polypogon monspeliensis
|
| 8685 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22549 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 8683 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 73 |
DnaJ protein |
321149989 |
F2VPR1_SOLNI |
Solanum nigrum
|
| 22695 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 8775 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 22739 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 16608 |
ATPase |
312075807 |
E1FVR9_LOALO |
Loa loa
|
| 8864 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 22722 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 22797 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 8824 |
ATPase |
359448103 |
|
Pseudoalteromonas sp. BSi20480
|
| 2935 |
ATP synthase F0 subunit 8 |
164652586 |
B2C9D6_TETUR |
Tetranychus urticae
|
| 22626 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 22981 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22963 |
ATPase, putative |
58266878 |
Q5KI67_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 22794 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 8890 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 8982 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 8671 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 2951 |
ATPase |
262301527 |
D0UKU0_ACHDO |
Acheta domesticus
|
| 8883 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 23031 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8990 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 2947 |
Putative nucleotide-binding protein |
269115406 |
D1MLM1_MAYDE |
Mayetiola destructor
|
| 22643 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8942 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 22507 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22888 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 22585 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 16667 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 22828 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8985 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 2959 |
ATP synthase F0 subunit 8 |
164652586 |
B2C9D6_TETUR |
Tetranychus urticae
|
| 75 |
ATP synthase subunit beta |
12004131 |
Q9GES6_ANAAR |
Anagallis arvensis
|
| 8674 |
ATPase |
359459786 |
|
Acaryochloris sp. CCMEE 5410
|
| 22659 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 22675 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22980 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 16587 |
ATPase |
312079317 |
E1G060_LOALO |
Loa loa
|
| 23034 |
ATPase, putative |
58264214 |
Q5KLI4_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 8862 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 8813 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8815 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 8708 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22627 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 8947 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 22645 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 22997 |
ATPase, putative |
58264774 |
Q5KKM6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 22755 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22810 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 8803 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8706 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 3544 |
ABC transporter family C protein ABCC2 |
336239515 |
G3JWY5_PLUXY |
Plutella xylostella
|
| 16752 |
ATPase |
312077420 |
E1FXT4_LOALO |
Loa loa
|
| 2940 |
Putative uncharacterized protein |
322780804 |
E9JAZ3_SOLIN |
Solenopsis invicta
|
| 22962 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22872 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 22999 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 2961 |
ATPase 6 |
186911146 |
B2L9C6_9MUSC |
Hydrellia tritici
|
| 8665 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 67 |
ATP synthase subunit beta |
110915694 |
A9L9Y6_FESAR |
Festuca arundinacea
|
| 2956 |
ATP synthase protein 8 |
12585179 |
ATP8_CERCA |
Ceratitis capitata
|
| 8958 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22921 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 2937 |
Vacuolar ATP synthase subunit B |
119874563 |
A8DRV1_9CRUS |
Triops longicaudatus
|
| 8726 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 22658 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 8846 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8819 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 22694 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 8816 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 3797 |
Plasma membrane calcium-transporting ATPase 3 |
332020943 |
F4WWU9_ACREC |
Acromyrmex echinatior
|
| 23012 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8991 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 8961 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 3617 |
PREDICTED: plasma membrane calcium-transporting ATPase 3-like isoform 1 |
328724399 |
|
Bellis perennis
|
| 8795 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22655 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 22925 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 90 |
ATP synthase subunit c, chloroplastic |
122225260 |
ATPH_HELAN |
Helianthus annuus
|
| 2960 |
Putative vacuolar ATP synthase subunit E |
299473919 |
D9J128_NILLU |
Nilaparvata lugens
|
| 22829 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 22959 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 2950 |
ATPase subunit 8 |
187370808 |
D7SGR8_9ACAR |
Panonychus ulmi
|
| 8843 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 8749 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 8963 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 8927 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22998 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8836 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22713 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 23028 |
ATPase, putative |
58271254 |
Q5KC30_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 8731 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22677 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22565 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 8871 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 93 |
ATP synthase subunit beta, chloroplastic |
75330483 |
ATPB_CONAR |
Convolvulus arvensis
|
| 22740 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 8771 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 8962 |
ATPase |
359459786 |
|
Acaryochloris sp. CCMEE 5410
|
| 8842 |
ATPase |
359456333 |
|
Pseudoalteromonas sp. BSi20495
|
| 8964 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 3875 |
ATPase beta subunit |
12666551 |
Q9BLQ1_MYZPE |
Myzus persicae
|
| 8945 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 8925 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 22958 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22924 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 22795 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 22547 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 22848 |
ATPase |
331250718 |
E3LAH1_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 23049 |
ATPase, putative |
58270374 |
Q5KCN0_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 22567 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 22607 |
ATPase, putative |
321253476 |
E6R3A7_CRYGW |
Cryptococcus gattii WM276
|
| 8710 |
ATPase |
359780641 |
|
Pseudomonas psychrotolerans L19
|
| 8778 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 22606 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 8703 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8940 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 16544 |
ATPase |
312086086 |
E1G877_LOALO |
Loa loa
|
| 3619 |
Vacuolar ATPase G subunit-like protein |
90820012 |
Q1W2A2_9HEMI |
Graphocephala atropunctata
|
| 84 |
ATP synthase subunit alpha |
57115589 |
Q5IBI9_PLALA |
Plantago lanceolata
|
| 22545 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 8983 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22544 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8669 |
ATPase |
359456333 |
|
Pseudoalteromonas sp. BSi20495
|
| 8909 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 23063 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8796 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22940 |
ATPase |
331220918 |
E3K3R3_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 16562 |
ATPase |
312086086 |
E1G877_LOALO |
Loa loa
|
| 22943 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 16565 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 8801 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 8712 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 22564 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 16543 |
ATPase |
339245927 |
E5SH67_TRISP |
Trichinella spiralis
|
| 8790 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 3550 |
Trehalose 6-phosphate synthase |
187234344 |
A4GHG0_SPOEX |
Spodoptera exigua
|
| 8692 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 23064 |
ATPase |
331234292 |
E3KM38_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 23050 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8690 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 22752 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8861 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 92 |
ATP synthase subunit alpha, chloroplastic |
290490184 |
D3WCJ2_PHOSE |
Phoradendron serotinum
|
| 8663 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 23032 |
ATPase, putative |
321264530 |
E6REU4_CRYGW |
Cryptococcus gattii WM276
|
| 8793 |
ATPase |
359686021 |
|
Leptospira santarosai str. 2000030832
|
| 8668 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
| 8980 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8781 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 8756 |
ATPase |
359449990 |
|
Pseudoalteromonas sp. BSi20480
|
| 55 |
ATP synthase subunit c, chloroplastic |
223634976 |
ATPH_IPOPU |
Ipomoea purpurea
|
| 8941 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 8705 |
Chaperone protein htpG |
16128457 |
HTPG_ECOLI |
Escherichia coli K-12
|
| 22588 |
Predicted ATPase |
169784291 |
Q2PIQ0_ASPOR |
Aspergillus oryzae RIB40
|
| 8812 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8848 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8774 |
ATPase |
359810463 |
|
Legionella drancourtii LLAP12
|
| 8818 |
ATPase |
359449990 |
|
Pseudoalteromonas sp. BSi20480
|
| 8684 |
ATPase |
359795777 |
|
Achromobacter sp. SY8
|
| 22889 |
ATPase, putative |
321257064 |
E6R5Q9_CRYGW |
Cryptococcus gattii WM276
|
| 22644 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22904 |
ATPase |
331231923 |
E3KIT0_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 8833 |
ATPase |
359683396 |
|
Leptospira santarosai str. 2000030832
|
| 8709 |
ATPase |
359795543 |
|
Achromobacter sp. SY8
|
| 16566 |
ATPase |
312082008 |
E1G3F4_LOALO |
Loa loa
|
| 22676 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 22548 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22979 |
ATPase |
331217656 |
E3JYG2_PUCGT |
Puccinia graminis f. sp. tritici CRL 75-36-700-3
|
| 2943 |
Putative nucleotide-binding protein |
269115406 |
D1MLM1_MAYDE |
Mayetiola destructor
|
| 22885 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 8727 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 8926 |
ATPase |
359726529 |
|
Leptospira weilii str. 2006001855
|
| 22814 |
ATPase |
150951383 |
A3GF11_PICST |
Scheffersomyces stipitis CBS 6054
|
| 8837 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 22756 |
ATPase, putative |
321248397 |
E6QZ17_CRYGW |
Cryptococcus gattii WM276
|
| 8887 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 16542 |
ATPase |
312077420 |
E1FXT4_LOALO |
Loa loa
|
| 22960 |
ATPase |
317034896 |
|
Aspergillus niger CBS 513.88
|
| 8978 |
ATPase |
359789638 |
|
Mesorhizobium alhagi CCNWXJ12-2
|
| 16668 |
ATPase |
312082008 |
E1G3F4_LOALO |
Loa loa
|
| 8868 |
ATPase |
359461011 |
|
Acaryochloris sp. CCMEE 5410
|
| 22977 |
ATPase, putative; Ino80p |
321262603 |
E6RBQ8_CRYGW |
Cryptococcus gattii WM276
|
| 22569 |
ATPase, putative |
321261730 |
E6RAE1_CRYGW |
Cryptococcus gattii WM276
|
| 22610 |
ATPase , putative |
321261567 |
E6R9S6_CRYGW |
Cryptococcus gattii WM276
|
| 8672 |
ATPase |
359795912 |
|
Achromobacter sp. SY8
|
| 8689 |
ATPase |
359781360 |
|
Pseudomonas psychrotolerans L19
|
| 8881 |
ATPase |
359783865 |
|
Pseudomonas psychrotolerans L19
|
 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;