anthraquinone
Identification (Source:EPA,EU)
| ID |
366 |
| Name |
anthraquinone |
| CAS |
84-65-1 |
| IUPAC |
anthraquinone |
| Inchi |
InChI=1S/C14H8O2/c15-13-9-5-1-2-6-10(9)14(16)12-8-4-3-7-11(12)13/h1-8H |
| CIPAC |
290 |
| EPA Code |
122701 |
| Formula |
C14H8O2 |
| SMILES |
O=C2c1ccccc1C(=O)c3ccccc23 |
| Status |
Not Approved |
Classification
| Mode of Action |
Induces retching in birds and so deters attack |
| Pesticide Type |
Repellent |
| Chemical Group |
Unclassified |
| Classification |
Bird repellents
|
Properties (Source:PubChem)
| Molecular Weight |
208.21 |
| Physical State |
Yellow to green crystals |
| AlogP |
2.808 |
| H-Bond Donor |
0 |
| H-Bond Acceptor |
2 |
| Surface Area |
188.84 |
| Polar Surface Area |
34.14 |
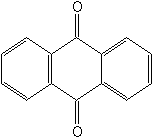
Structure
Ecotoxicology (Source:PPDB,IPM)
| Species |
Method |
Study Time |
Dose Type |
Value |
| Algae |
Acute |
72 hour |
EC50, growth |
10 mg l-1 |
| Aquatic crustaceans |
Acute |
96 hour |
LC50 |
0.942 mg l-1 |
| Aquatic invertebrates |
Acute |
48 hour |
EC50 |
10 mg l-1 |
| Aquatic plants |
Acute |
7 day |
EC50, biomass |
0.2 mg l-1 |
| Birds |
Acute |
|
LD50 |
2000 mg kg-1 |
| Earthworms |
Acute |
14 day |
LD50 |
1000 mg kg-1 |
| Fish |
Acute |
96 hour |
LC50 |
72 mg l-1 |
| Mammals |
Acute oral |
|
LD50 |
> 5000 mg kg-1 |
| Mammals |
Short term dietary |
|
NOEL |
15 ppm diet |
|
Bio-concentration factor |
|
CT50 |
Not available days |
|
Bio-concentration factor |
|
BCF |
100 |
Related targets
| Target Name |
RScore (About this table) |
|
|
Telomerase (protein family or complex)
|
428(4,6,7,43) |
Details |
|
luteinizing hormone releasing hormone
|
373(4,5,5,23) |
Details |
|
casein kinase II
|
331(4,4,4,11) |
Details |
|
cytochrome c
|
342(3,5,6,37) |
Details |
|
ICRF
|
296(3,4,4,26) |
Details |
|
albumin
|
252(3,3,3,12) |
Details |
|
p56lck
|
248(3,3,3,8) |
Details |
|
Prolidase
|
230(2,4,4,10) |
Details |
|
lysozyme
|
212(2,3,5,12) |
Details |
|
aquaporin 4
|
191(2,2,5,16) |
Details |
|
NMDA receptors (protein family or complex)
|
189(2,2,6,9) |
Details |
|
NADH dehydrogenase
|
188(2,2,5,13) |
Details |
|
DT diaphorase
|
187(2,2,3,22) |
Details |
|
NADPH cytochrome P450 reductase
|
185(2,2,4,15) |
Details |
|
C18
|
182(2,2,3,17) |
Details |
|
protein kinase C (protein family or complex)
|
175(2,2,2,15) |
Details |
|
caspases (protein family or complex)
|
173(2,2,2,13) |
Details |
|
alcohol dehydrogenase (protein family or complex)
|
168(2,2,3,3) |
Details |
|
activator protein 1
|
167(2,2,2,7) |
Details |
|
glucose 6 phosphate translocase
|
166(2,2,2,6) |
Details |
|
NTPDase3
|
164(2,2,2,4) |
Details |
|
erythrocyte AMP deaminase
|
162(2,2,2,2) |
Details |
|
calmodulin
|
156(1,3,3,16) |
Details |
|
ferritin (protein family or complex)
|
116(1,2,2,6) |
Details |
|
protein tyrosine kinase
|
115(1,2,2,5) |
Details |
|
cathepsin B
|
114(1,2,2,4) |
Details |
|
RNase
|
113(1,2,2,3) |
Details |
|
Dec 1
|
111(1,1,5,11) |
Details |
|
xanthine oxidase
|
100(1,1,3,10) |
Details |
|
alkaline phosphatase
|
100(1,1,2,15) |
Details |
|
aquaporin 2
|
100(1,1,3,10) |
Details |
|
ERK1
|
97(1,1,1,17) |
Details |
|
ATPase
|
91(1,1,1,11) |
Details |
|
phosphodiesterase
|
91(1,1,2,6) |
Details |
|
protein kinase A (protein family or complex)
|
91(1,1,1,11) |
Details |
|
P gp
|
90(1,1,1,10) |
Details |
|
interferon (protein family or complex)
|
89(1,1,1,9) |
Details |
|
hexokinase
|
89(1,1,1,9) |
Details |
|
lactate dehydrogenase (protein family or complex)
|
88(1,1,1,8) |
Details |
|
telomerase reverse transcriptase
|
87(1,1,1,7) |
Details |
|
caspase 8
|
86(1,1,1,6) |
Details |
|
CA1
|
86(1,1,1,6) |
Details |
|
tumor necrosis factor
|
86(1,1,1,6) |
Details |
|
NA K ATPase
|
84(1,1,1,4) |
Details |
|
S protein
|
84(1,1,1,4) |
Details |
|
malate dehydrogenase
|
84(1,1,1,4) |
Details |
|
lactoperoxidase
|
84(1,1,1,4) |
Details |
|
phosphoglycerate kinase
|
84(1,1,1,4) |
Details |
|
angiotensin converting enzyme 2
|
84(1,1,1,4) |
Details |
|
C13
|
83(1,1,1,3) |
Details |
|
PC3
|
83(1,1,1,3) |
Details |
|
P2X1
|
82(1,1,1,2) |
Details |
|
beta hexosaminidase
|
82(1,1,1,2) |
Details |
|
P2X receptors (protein family or complex)
|
82(1,1,1,2) |
Details |
|
KNO
|
81(1,1,1,1) |
Details |
|
Tyrosinase
|
81(1,1,1,1) |
Details |
|
matrix metalloproteinase 1
|
81(1,1,1,1) |
Details |
|
glutamate receptors (protein family or complex)
|
81(1,1,1,1) |
Details |
|
serine threonine protein kinases
|
81(1,1,1,1) |
Details |
|
MutS
|
81(1,1,1,1) |
Details |
|
cytochrome P450 (protein family or complex)
|
132(0,3,4,37) |
Details |
|
complex is
|
105(0,3,3,15) |
Details |
|
superoxide dismutase
|
88(0,2,3,23) |
Details |
|
aryl hydrocarbon receptor
|
86(0,2,2,26) |
Details |
|
Red 2
|
85(0,2,5,10) |
Details |
|
interleukin 1
|
76(0,2,2,16) |
Details |
|
ring 1
|
74(0,2,4,4) |
Details |
|
AP 1 (protein family or complex)
|
71(0,2,2,11) |
Details |
|
DCIP
|
70(0,2,3,5) |
Details |
|
cyclic nucleotide phosphodiesterase
|
69(0,2,2,9) |
Details |
|
RR2
|
57(0,1,3,17) |
Details |
|
caspase 3
|
46(0,1,1,16) |
Details |
|
S12
|
46(0,1,3,6) |
Details |
|
ASAM
|
46(0,1,3,6) |
Details |
|
MR 1
|
44(0,1,1,14) |
Details |
|
catalase
|
40(0,1,1,10) |
Details |
|
estrogen receptor
|
40(0,1,1,10) |
Details |
|
plasma hyaluronan binding protein
|
39(0,1,1,9) |
Details |
|
myoglobin
|
39(0,1,1,9) |
Details |
|
BLT2
|
39(0,1,1,9) |
Details |
|
pg m
|
38(0,1,2,3) |
Details |
|
casein
|
36(0,1,1,6) |
Details |
|
NR1
|
35(0,1,1,5) |
Details |
|
fibronectin
|
34(0,1,1,4) |
Details |
|
protein is
|
34(0,1,1,4) |
Details |
|
HEMA
|
34(0,1,1,4) |
Details |
|
PLA2
|
34(0,1,1,4) |
Details |
|
H ras
|
34(0,1,1,4) |
Details |
|
CNSN
|
34(0,1,1,4) |
Details |
|
phosphomannomutase
|
33(0,1,1,3) |
Details |
|
P2Y1
|
32(0,1,1,2) |
Details |
|
ACAC
|
31(0,1,1,1) |
Details |
|
protein phosphatase 2A (protein family or complex)
|
31(0,1,1,1) |
Details |
|
IgE
|
31(0,1,1,1) |
Details |
|
P21
|
31(0,1,1,1) |
Details |
|
lipase
|
31(0,1,1,1) |
Details |
|
cholinesterase
|
31(0,1,1,1) |
Details |
|
lag 5
|
31(0,1,1,1) |
Details |
|
APT2
|
31(0,1,1,1) |
Details |
|
5 lipoxygenase
|
31(0,1,1,1) |
Details |
|
quinone reductase
|
36(0,0,2,26) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;