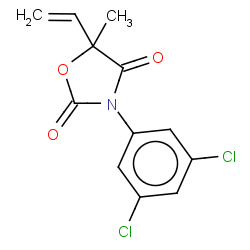
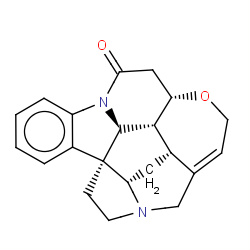
| 2815 |
Putative uncharacterized protein |
270011756 |
D6WW14_TRICA |
Tribolium castaneum
|
| 2819 |
Chemosensory protein |
159162570 |
Q9NG96_MAMBR |
Mamestra brassicae
|
| 575 |
Photosystem II reaction center protein I |
212277646 |
PSBI_IPOPU |
Ipomoea purpurea
|
| 2813 |
White eye protein |
23953881 |
Q8IS30_BACCU |
Bactrocera cucurbitae
|
| 2812 |
Putative uncharacterized protein |
322780411 |
E9JB70_SOLIN |
Solenopsis invicta
|
| 572 |
Photosystem II reaction center protein I |
212277649 |
PSBI_LOLPR |
Lolium perenne
|
| 2824 |
Endo-polygalacturonase |
3122616 |
PGLR_DIAAB |
Diaprepes abbreviatus
|
| 2818 |
Putative defense protein Hdd11 |
74873244 |
DFP11_HYPCU |
Hyphantria cunea
|
| 2821 |
Adenosine deaminase AGSA |
325297112 |
AGSA_APLCA |
Aplysia californica
|
| 2820 |
GD18869 |
194199559 |
B4R1Y3_DROSI |
Drosophila simulans
|
| 2808 |
Putative potassium antiporter CHAC |
204307480 |
D2CSM0_HOMVI |
Homalodisca vitripennis
|
| 2814 |
Heterogeneous nuclear ribonucleoprotein A1, A2/B1 homolog |
133256 |
ROA1_SCHAM |
Schistocerca americana
|
| 2817 |
Ubiquitin-40S ribosomal protein S27a |
302393750 |
RS27A_MANSE |
Manduca sexta
|
| 3802 |
Putative uncharacterized protein |
322780411 |
E9JB70_SOLIN |
Solenopsis invicta
|
| 2825 |
Ubiquitin-40S ribosomal protein S27a |
302393756 |
RS27A_PLUXY |
Plutella xylostella
|
| 576 |
Cytochrome c oxidase subunit 5C-2 |
48428106 |
CX5C2_HELAN |
Helianthus annuus
|
| 568 |
Photosystem II reaction center protein J |
150413786 |
PSBJ_CAPBU |
Capsella bursa-pastoris
|
| 571 |
Ubiquitin-60S ribosomal protein L40 |
302393709 |
RL40_BRARA |
Brassica rapa subsp. rapa
|
| 566 |
Cytochrome c oxidase subunit 5C |
117107 |
COX5C_IPOBA |
Ipomoea batatas
|
| 2823 |
Possible lysine-specific histone demethylase 1 |
75027620 |
LSDA_DROME |
Drosophila melanogaster
|
| 2810 |
Glycine dehydrogenase [decarboxylating], mitochondrial |
121083 |
GCSP_PEA |
Pisum sativum
|
| 573 |
Chloroplast chlorophyll a/b-binding protein |
295824591 |
B3RFS0_SOLNI |
Solanum nigrum
|
| 2816 |
Ribosomal protein S28e-like protein |
121543975 |
A2I483_MACHI |
Maconellicoccus hirsutus
|
| 2822 |
60S acidic ribosomal protein P0 |
20139848 |
RLA0_CERCA |
Ceratitis capitata
|
| 567 |
HMG1/2-like protein |
729736 |
HMGL_IPONI |
Ipomoea nil
|
| 570 |
Photosystem II reaction center protein I |
125980841 |
PSBI_AGRST |
Agrostis stolonifera
|
| 2827 |
PREDICTED: GTPase Era-like |
328725521 |
|
Sternorrhyncha
|
| 2811 |
p77 homologue |
305632939 |
E0D3H4_SPOLT |
Spodoptera litura
|
| 2809 |
Azurocidin-like protein |
21311448 |
Q8MVZ0_TRINI |
Trichoplusia ni
|
| 569 |
Cytochrome b6-f complex iron-sulfur subunit, chloroplastic |
68566191 |
UCRIA_WHEAT |
Triticum aestivum
|
| 577 |
Photosystem II reaction center protein J |
62287059 |
PSBJ_HORJU |
Hordeum jubatum
|
| 574 |
Photosystem I P700 chlorophyll a apoprotein A2 |
122219010 |
PSAB_CUSSA |
Cuscuta sandwichiana
|
| 2826 |
Ribosomal protein S28e-like protein |
121543975 |
A2I483_MACHI |
Maconellicoccus hirsutus
|
| 837 |
Major pollen allergen Dac g 4 |
32363463 |
MPAG4_DACGL |
Dactylis glomerata
|
 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;