azobenzene
Identification (Source:EPA,EU)
| ID |
106 |
| Name |
azobenzene |
| CAS |
103-33-3 |
| IUPAC |
azobenzene |
| Inchi |
InChI=1S/C12H10N2/c1-3-7-11(8-4-1)13-14-12-9-5-2-6-10-12/h1-10H |
| CIPAC |
241 |
| EPA Code |
007401 |
| Formula |
C12H10N2 |
| SMILES |
N(=N/c1ccccc1)\c2ccccc2 |
| Status |
Not Available |
Classification
| Mode of Action |
Acts by inhibiting oxidative phosphorylation. |
| Pesticide Type |
Acaricide, Ovicide, Miticide |
| Chemical Group |
Bridged diphenyl |
| Classification |
bridged diphenyl acaricides
|
Properties (Source:PubChem)
| Molecular Weight |
182.22 |
| Physical State |
Orange crystals |
| AlogP |
4.199 |
| H-Bond Donor |
0 |
| H-Bond Acceptor |
2 |
| Surface Area |
180.12 |
| Polar Surface Area |
24.72 |
Structure
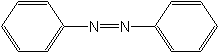
Ecotoxicology (Source:PPDB,IPM)
| Species |
Method |
Study Time |
Dose Type |
Value |
| Algae |
Acute |
72 hour |
EC50, growth |
1.7 mg l-1 |
| Aquatic invertebrates |
Acute |
48 hour |
EC50 |
5.0 mg l-1 |
| Aquatic invertebrates |
Chronic |
21 day |
NOEC |
0.009 mg l-1 |
| Fish |
Acute |
96 hour |
LC50 |
0.5 mg l-1 |
| Mammals |
Acute oral |
|
LD50 |
> 1000 mg kg-1 |
Related targets
| Target Name |
RScore (About this table) |
|
|
acetylcholine receptor (protein family or complex)
|
240(2,4,4,20) |
Details |
|
ATPase
|
201(2,3,3,11) |
Details |
|
thrombin
|
173(2,2,2,13) |
Details |
|
CDSP
|
164(2,2,2,4) |
Details |
|
Red 1
|
150(1,3,3,10) |
Details |
|
kinesin
|
145(1,3,3,5) |
Details |
|
PGDH
|
116(1,2,2,6) |
Details |
|
SANS
|
106(1,1,3,16) |
Details |
|
complex is
|
95(1,1,1,15) |
Details |
|
SP6
|
90(1,1,2,5) |
Details |
|
Ah receptor
|
89(1,1,1,9) |
Details |
|
HEMA
|
89(1,1,2,4) |
Details |
|
monoamine oxidase
|
87(1,1,2,2) |
Details |
|
alpha fetoprotein
|
84(1,1,1,4) |
Details |
|
albumin
|
84(1,1,1,4) |
Details |
|
Tyrosinase
|
83(1,1,1,3) |
Details |
|
C18
|
83(1,1,1,3) |
Details |
|
hexokinase
|
83(1,1,1,3) |
Details |
|
Myosin
|
83(1,1,1,3) |
Details |
|
transferrin
|
82(1,1,1,2) |
Details |
|
phospholipase A2
|
82(1,1,1,2) |
Details |
|
calmodulin
|
82(1,1,1,2) |
Details |
|
Rhodanese
|
82(1,1,1,2) |
Details |
|
SERS
|
99(0,2,6,19) |
Details |
|
DR 1
|
64(0,2,2,4) |
Details |
|
cytochrome P450 (protein family or complex)
|
52(0,1,2,17) |
Details |
|
Bcl xL
|
36(0,1,1,6) |
Details |
|
thioredoxin reductase
|
34(0,1,1,4) |
Details |
|
glutamate receptor
|
34(0,1,1,4) |
Details |
|
glutathione S transferase
|
34(0,1,1,4) |
Details |
|
brains
|
33(0,1,1,3) |
Details |
|
ADCA
|
33(0,1,1,3) |
Details |
|
PCCA
|
32(0,1,1,2) |
Details |
|
ClN 3
|
32(0,1,1,2) |
Details |
|
chaperonin
|
32(0,1,1,2) |
Details |
|
hemolysin
|
32(0,1,1,2) |
Details |
|
aldolase A
|
32(0,1,1,2) |
Details |
|
H ras
|
32(0,1,1,2) |
Details |
|
phosphodiesterase
|
32(0,1,1,2) |
Details |
|
Ho 1
|
31(0,1,1,1) |
Details |
|
CDs 1
|
31(0,1,1,1) |
Details |
|
NATI
|
31(0,1,1,1) |
Details |
|
protein disulfide isomerase
|
31(0,1,1,1) |
Details |
|
ps 2
|
31(0,1,1,1) |
Details |
|
RNase
|
19(0,0,3,4) |
Details |
|
catalase
|
15(0,0,2,5) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;
 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;