Hsp70
General Information
| Text mining term | Hsp70 |
|---|---|
| Synonym | APG 2; APG2; HS24/P52; HSP70RY; HSPA 4; HSPA4; Heat shock 70 kDa protein 4; Heat shock 70 related protein APG 2… |
| Relative Score | 440(3,5,12,105) |
| Total Confidence | 0.16 |
Sequence (Source: NCBI Gene,UniProt)
Related Compounds
| Compound Name | Structure | RScore(About this table) | |
|---|---|---|---|
| dichlorvos |
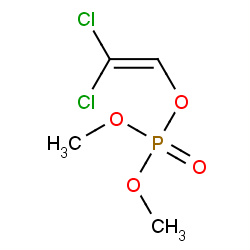
|
166(2,2,2,6) | Details |
| paraquat |

|
87(1,1,1,7) | Details |
| cycloheximide |
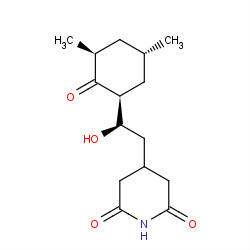
|
54(0,1,1,24) | Details |
| 2,4-D |

|
39(0,1,2,4) | Details |
| pentachlorophenol |

|
27(0,0,2,17) | Details |
| cypermethrin |

|
14(0,0,2,4) | Details |
| mercuric chloride |
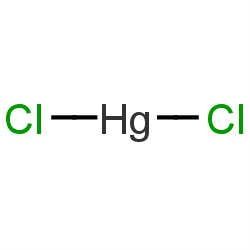
|
12(0,0,0,12) | Details |
| ethephon |

|
8(0,0,1,3) | Details |
| chlorpyrifos |
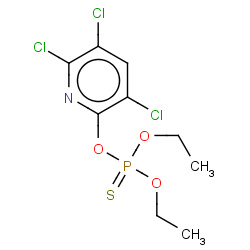
|
7(0,0,1,2) | Details |
| SMA |

|
5(0,0,0,5) | Details |
| sodium azide |

|
5(0,0,0,5) | Details |
| pyrethrins |
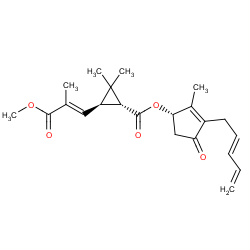
|
4(0,0,0,4) | Details |
| rotenone |
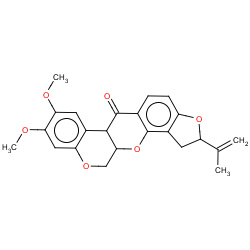
|
3(0,0,0,3) | Details |
| copper arsenate |

|
2(0,0,0,2) | Details |
| parathion |
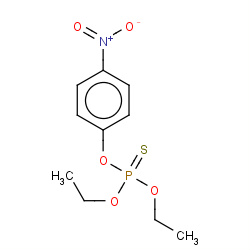
|
2(0,0,0,2) | Details |
| vinclozolin |
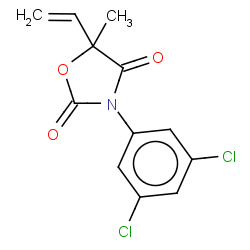
|
1(0,0,0,1) | Details |
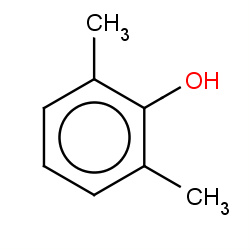
| xylenols |
|
1(0,0,0,1) | Details |
 Network initializing...Please wait
Network initializing...Please wait
Node List
| ID | Name | Type |
|---|---|---|
Legend:
 : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;