| 24701 |
phospholipase C |
336262013 |
|
Sordaria macrospora k-hell
|
| 289 |
Phosphoinositide-specific phospholipase C |
4588474 |
Q9XEK4_BRANA |
Brassica napus
|
| 24773 |
Phospholipase C, putative |
58259579 |
Q5KNT6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 24699 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 24838 |
Phospholipase C |
296805908 |
C5FYZ4_ARTOC |
Arthroderma otae CBS 113480
|
| 24702 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 24746 |
phospholipase C |
336262013 |
|
Sordaria macrospora k-hell
|
| 24793 |
Phospholipase C |
261203269 |
C5JC29_AJEDS |
Ajellomyces dermatitidis SLH14081
|
| 24841 |
Phospholipase C |
261203269 |
C5JC29_AJEDS |
Ajellomyces dermatitidis SLH14081
|
| 2114 |
Phospholipase C |
325296761 |
Q27J08_APLCA |
Aplysia californica
|
| 24794 |
phospholipase C |
336262013 |
|
Sordaria macrospora k-hell
|
| 24769 |
Phospholipase C |
299738283 |
A8P234_COPC7 |
Coprinopsis cinerea okayama7#130
|
| 24700 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 24724 |
phospholipase C |
336262013 |
|
Sordaria macrospora k-hell
|
| 24817 |
Phospholipase C |
299738283 |
A8P234_COPC7 |
Coprinopsis cinerea okayama7#130
|
| 2116 |
PREDICTED: 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase-like isoform 2 |
328724636 |
|
Sisymbrium irio
|
| 2108 |
PREDICTED: 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase-like isoform 2 |
328724636 |
|
Bellis perennis
|
| 286 |
Phosphoinositide-specific phospholipase C |
10880265 |
Q9FSW1_9POAL |
Digitaria sanguinalis
|
| 24842 |
phospholipase C |
336262013 |
|
Sordaria macrospora k-hell
|
| 2112 |
Phosphatidylinositol-specific phospholipase C X domain-containing protein 1 |
307207209 |
E2BH78_HARSA |
Harpegnathos saltator
|
| 24795 |
Phospholipase C |
296805908 |
C5FYZ4_ARTOC |
Arthroderma otae CBS 113480
|
| 288 |
Phosphoinositide-specific phospholipase C |
10880265 |
Q9FSW1_9POAL |
Digitaria sanguinalis
|
| 2115 |
No receptor potential A, isoform E |
272505967 |
E1JJD5_DROME |
Drosophila melanogaster
|
| 24816 |
Phospholipase C |
254574064 |
C4R8N7_PICPG |
Pichia pastoris GS115
|
| 2117 |
PREDICTED: 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase-like isoform 2 |
328724636 |
|
Sternorrhyncha
|
| 24747 |
Phospholipase C |
299738283 |
A8P234_COPC7 |
Coprinopsis cinerea okayama7#130
|
| 24745 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 24748 |
Phospholipase C, putative |
58259579 |
Q5KNT6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 24725 |
Phospholipase C, putative |
58259579 |
Q5KNT6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 2109 |
Phospholipase C |
22085144 |
Q8LLW1_PEA |
Pisum sativum
|
| 24770 |
Phospholipase C |
296805908 |
C5FYZ4_ARTOC |
Arthroderma otae CBS 113480
|
| 24771 |
Phospholipase C, putative |
58259579 |
Q5KNT6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|
| 24814 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 2113 |
GD23017 |
194189633 |
B4Q625_DROSI |
Drosophila simulans
|
| 2111 |
PREDICTED: similar to phospholipase c beta isoform 3 |
189236413 |
|
Soliva sessilis
|
| 2110 |
Putative uncharacterized protein |
322796572 |
E9IK32_SOLIN |
Solenopsis invicta
|
| 24722 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 24792 |
Phospholipase C |
299738283 |
A8P234_COPC7 |
Coprinopsis cinerea okayama7#130
|
| 287 |
Phosphoinositide-specific phospholipase C2 |
312618320 |
E5G604_WHEAT |
Triticum aestivum
|
| 24772 |
Phospholipase C, putative |
321249742 |
E6QY51_CRYGW |
Cryptococcus gattii WM276
|
| 24723 |
phospholipase C |
336262013 |
|
Sordaria macrospora k-hell
|
| 24815 |
Phospholipase C, putative |
58259579 |
Q5KNT6_CRYNJ |
Cryptococcus neoformans var. neoformans JEC21
|


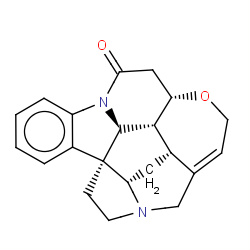
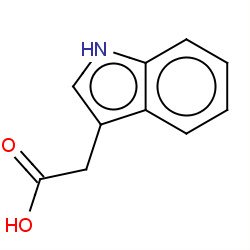
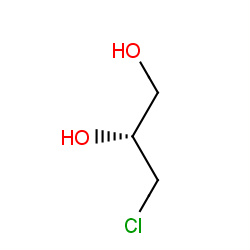

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;