4-aminopyridine
Identification (Source:EPA,EU)
| ID |
332 |
| Name |
4-aminopyridine |
| CAS |
504-24-5 |
| IUPAC |
pyridin-4-amine |
| Inchi |
InChI=1S/C5H6N2/c6-5-1-3-7-4-2-5/h1-4H,(H2,6,7) |
| CIPAC |
None allocated |
| EPA Code |
069201 |
| Formula |
C5H6N2 |
| SMILES |
c1(ccncc1)N |
| Status |
Not Available |
Classification
| Mode of Action |
Works as a potassium channel blocker, over stimulating the nervous system |
| Pesticide Type |
Avicide, Acaricide, Repellent |
| Chemical Group |
Isomeric amine of pyridine |
| Classification |
Avicides
|
Properties (Source:PubChem)
| Molecular Weight |
94.13 |
| Physical State |
White crystalline solid |
| AlogP |
-0.068 |
| H-Bond Donor |
1 |
| H-Bond Acceptor |
2 |
| Surface Area |
104.31 |
| Polar Surface Area |
38.9 |
Structure
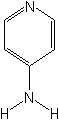
Ecotoxicology (Source:PPDB,IPM)
| Species |
Method |
Study Time |
Dose Type |
Value |
| Aquatic invertebrates |
Acute |
48 hour |
EC50 |
3.2 mg l-1 |
| Birds |
Acute |
|
LD50 |
722 mg kg-1 |
| Fish |
Acute |
96 hour |
LC50 |
3.4 mg l-1 |
| Mammals |
Acute oral |
|
LD50 |
29 mg kg-1 |
Related targets
| Target Name |
RScore (About this table) |
|
|
Potassium channel (protein family or complex)
|
2571(5,60,110,271) |
Details |
|
CA3
|
805(5,11,27,145) |
Details |
|
protein kinase C (protein family or complex)
|
475(4,7,7,65) |
Details |
|
NMDA receptors (protein family or complex)
|
546(3,10,11,91) |
Details |
|
glutamate receptor
|
361(2,7,7,51) |
Details |
|
angiotensin II
|
280(2,4,4,60) |
Details |
|
GABA B receptors
|
233(2,3,3,43) |
Details |
|
glutamate receptors (protein family or complex)
|
201(2,2,3,36) |
Details |
|
episodic ataxia type 2
|
189(2,2,4,19) |
Details |
|
voltage gated K channel (protein family or complex)
|
179(2,2,3,14) |
Details |
|
parvalbumin
|
177(2,2,3,12) |
Details |
|
cholinesterase
|
169(2,2,2,9) |
Details |
|
epidermal growth factor receptor
|
166(2,2,2,6) |
Details |
|
CA1
|
520(1,8,17,185) |
Details |
|
voltage gated potassium channel
|
385(1,8,14,65) |
Details |
|
beta 3 adrenoceptors
|
227(1,5,6,22) |
Details |
|
muscles
|
273(1,4,7,88) |
Details |
|
AP 2
|
181(1,3,8,16) |
Details |
|
caspase (protein family or complex)
|
151(1,3,3,11) |
Details |
|
estrogen receptor
|
148(1,3,3,8) |
Details |
|
KCNA5
|
142(1,2,4,22) |
Details |
|
cAMP dependent protein kinase (protein family or complex)
|
142(1,2,3,27) |
Details |
|
beta adrenoceptors (protein family or complex)
|
138(1,2,2,28) |
Details |
|
SNAP
|
134(1,2,3,19) |
Details |
|
adenylate cyclase
|
118(1,2,2,8) |
Details |
|
purinergic receptor
|
112(1,2,2,2) |
Details |
|
KChIP2
|
113(1,1,2,28) |
Details |
|
Substance P
|
103(1,1,2,18) |
Details |
|
TREK 1
|
103(1,1,2,18) |
Details |
|
potassium ion channels
|
103(1,1,3,13) |
Details |
|
leptin
|
100(1,1,2,15) |
Details |
|
phospholipase C
|
99(1,1,2,14) |
Details |
|
mitogen activated protein kinase (protein family or complex)
|
99(1,1,1,19) |
Details |
|
catalase
|
96(1,1,1,16) |
Details |
|
pituitary adenylate cyclase activating polypeptide
|
95(1,1,2,10) |
Details |
|
olfactory receptor
|
92(1,1,1,12) |
Details |
|
IL 1beta
|
91(1,1,1,11) |
Details |
|
dopamine transporter
|
91(1,1,2,6) |
Details |
|
c kit
|
88(1,1,1,8) |
Details |
|
mcl 1
|
88(1,1,1,8) |
Details |
|
TRPV1
|
86(1,1,1,6) |
Details |
|
hASCs
|
86(1,1,1,6) |
Details |
|
integrin
|
85(1,1,1,5) |
Details |
|
KCNA1
|
85(1,1,1,5) |
Details |
|
gephyrin
|
85(1,1,1,5) |
Details |
|
aquaporin 1
|
84(1,1,1,4) |
Details |
|
HK2
|
83(1,1,1,3) |
Details |
|
interferon gamma
|
83(1,1,1,3) |
Details |
|
TRIM
|
82(1,1,1,2) |
Details |
|
alpha2 adrenoceptor (protein family or complex)
|
82(1,1,1,2) |
Details |
|
brain 4
|
82(1,1,1,2) |
Details |
|
adenosine kinase
|
82(1,1,1,2) |
Details |
|
laminin (protein family or complex)
|
82(1,1,1,2) |
Details |
|
proline rich tyrosine kinase 2
|
81(1,1,1,1) |
Details |
|
MAP4
|
81(1,1,1,1) |
Details |
|
AP 1 (protein family or complex)
|
373(0,11,15,23) |
Details |
|
calcium channel (protein family or complex)
|
206(0,5,6,51) |
Details |
|
guanylate cyclase
|
176(0,5,5,26) |
Details |
|
Acetylcholinesterase
|
111(0,3,3,21) |
Details |
|
adenosine A1 receptor
|
97(0,3,3,7) |
Details |
|
c fos
|
120(0,2,7,35) |
Details |
|
G protein
|
100(0,2,2,40) |
Details |
|
CB1 receptors
|
90(0,2,2,30) |
Details |
|
KCa1.1
|
89(0,2,2,29) |
Details |
|
AP 4
|
87(0,2,4,17) |
Details |
|
PACAP
|
87(0,2,2,27) |
Details |
|
mu opioid receptors
|
85(0,2,2,25) |
Details |
|
adenylyl cyclase
|
76(0,2,2,16) |
Details |
|
neuropeptide Y
|
76(0,2,3,11) |
Details |
|
GABA receptor (protein family or complex)
|
75(0,2,2,15) |
Details |
|
cytochrome P450 (protein family or complex)
|
69(0,2,2,9) |
Details |
|
TASK
|
84(0,1,1,54) |
Details |
|
ATPase
|
64(0,1,2,29) |
Details |
|
kininogen
|
55(0,1,2,20) |
Details |
|
endothelin 1
|
54(0,1,2,19) |
Details |
|
ERK1
|
53(0,1,2,18) |
Details |
|
sodium channel (protein family or complex)
|
51(0,1,1,21) |
Details |
|
AP 3 (protein family or complex)
|
50(0,1,3,10) |
Details |
|
mGluR5
|
47(0,1,1,17) |
Details |
|
glycine receptors (protein family or complex)
|
41(0,1,2,6) |
Details |
|
P13
|
41(0,1,2,6) |
Details |
|
complex I
|
40(0,1,1,10) |
Details |
|
bcl 2
|
39(0,1,1,9) |
Details |
|
SAP97
|
39(0,1,1,9) |
Details |
|
PP2
|
38(0,1,1,8) |
Details |
|
homer1
|
37(0,1,1,7) |
Details |
|
aquaporin 4
|
37(0,1,1,7) |
Details |
|
ET A
|
36(0,1,1,6) |
Details |
|
SCLC
|
36(0,1,1,6) |
Details |
|
TRPV2
|
36(0,1,1,6) |
Details |
|
Ach 1
|
35(0,1,1,5) |
Details |
|
NPPB
|
35(0,1,1,5) |
Details |
|
TRAM
|
35(0,1,1,5) |
Details |
|
GABA transporter
|
35(0,1,1,5) |
Details |
|
SUR2
|
35(0,1,1,5) |
Details |
|
cytochrome c
|
35(0,1,1,5) |
Details |
|
NHE1
|
34(0,1,1,4) |
Details |
|
TRPC6
|
34(0,1,1,4) |
Details |
|
FB1
|
34(0,1,1,4) |
Details |
|
myelin basic protein
|
34(0,1,1,4) |
Details |
|
eag
|
33(0,1,1,3) |
Details |
|
c Jun N terminal kinase (protein family or complex)
|
33(0,1,1,3) |
Details |
|
modulatory calcineurin interacting proteins
|
33(0,1,1,3) |
Details |
|
PDE3
|
33(0,1,1,3) |
Details |
|
GABA at
|
33(0,1,1,3) |
Details |
|
AT2
|
33(0,1,1,3) |
Details |
|
ERK2
|
32(0,1,1,2) |
Details |
|
P23
|
32(0,1,1,2) |
Details |
|
integrator (protein family or complex)
|
32(0,1,1,2) |
Details |
|
activated protein C
|
31(0,1,1,1) |
Details |
|
SOD1
|
31(0,1,1,1) |
Details |
|
ATP 3
|
31(0,1,1,1) |
Details |
|
CYP2C9
|
31(0,1,1,1) |
Details |
|
somatostatin
|
30(0,0,1,25) |
Details |
|
Cx36
|
26(0,0,2,16) |
Details |
|
neurotrophin
|
24(0,0,0,24) |
Details |
|
calcitonin gene related peptide
|
24(0,0,0,24) |
Details |
|
acetylcholine receptor (protein family or complex)
|
20(0,0,0,20) |
Details |
|
TRESK 2
|
18(0,0,1,13) |
Details |
|
phosphodiesterase
|
17(0,0,0,17) |
Details |
|
TASK3
|
16(0,0,0,16) |
Details |
|
KCa3.1
|
15(0,0,0,15) |
Details |
|
OBCAM
|
15(0,0,1,10) |
Details |
|
HM 1
|
15(0,0,1,10) |
Details |
|
5 HT2A
|
14(0,0,0,14) |
Details |
|
G alpha q
|
14(0,0,1,9) |
Details |
|
TASK 2
|
13(0,0,0,13) |
Details |
|
Ca v
|
13(0,0,0,13) |
Details |
|
SCA1
|
13(0,0,1,8) |
Details |
|
calmodulin
|
13(0,0,0,13) |
Details |
|
SIVA
|
13(0,0,2,3) |
Details |
|
MKL1
|
12(0,0,0,12) |
Details |
|
KCNQ1
|
12(0,0,0,12) |
Details |
|
p38
|
12(0,0,0,12) |
Details |
|
mGluR1
|
12(0,0,0,12) |
Details |
|
Rho kinase
|
11(0,0,0,11) |
Details |
|
RAPSN
|
11(0,0,0,11) |
Details |
|
NF2
|
11(0,0,1,6) |
Details |
|
protein phosphatase 2B (protein family or complex)
|
11(0,0,0,11) |
Details |
|
SK3
|
11(0,0,0,11) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;