kinetin
Identification (Source:EPA,EU)
| ID |
1700 |
| Name |
kinetin |
| CAS |
525-79-1 |
| IUPAC |
|
| Inchi |
InChI=1S/C10H9N5O/c1-2-7(16-3-1)4-11-9-8-10(13-5-12-8)15-6-14-9/h1-3,5-6H,4H2,(H2,11,12,13,14,15) |
| CIPAC |
|
| EPA Code |
116801 |
| Formula |
C10H9N5O |
| SMILES |
c12c(NCc3ccco3)ncnc1nc[nH]2 |
| Status |
Not Available |
Classification
| Mode of Action |
Substance promotes cell division, leaf expansion and slows the leaf aging processes |
| Pesticide Type |
Plant growth regulator, Nematicide |
| Chemical Group |
Plant derived |
| Classification |
cytokinins
|
Properties (Source:PubChem)
| Molecular Weight |
215.22 |
| Physical State |
White crystalline solid |
| AlogP |
0.403 |
| H-Bond Donor |
2 |
| H-Bond Acceptor |
4 |
| Surface Area |
206.95 |
| Polar Surface Area |
79.63 |
Structure
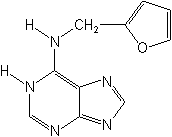
Ecotoxicology (Source:PPDB,IPM)
| Species |
Method |
Study Time |
Dose Type |
Value |
| Aquatic invertebrates |
Acute |
48 hour |
EC50 |
> 1000 mg l-1 |
| Fish |
Acute |
96 hour |
LC50 |
> 1000 mg l-1 |
| Mammals |
Acute oral |
|
LD50 |
> 5000 mg kg-1 |
| Mammals |
Acute oral |
|
LD50 |
> 5000 mg kg-1 |
Related targets
| Target Name |
RScore (About this table) |
|
|
CCND2
|
252(3,3,4,7) |
Details |
|
superoxide dismutase
|
206(2,3,3,16) |
Details |
|
xanthine oxidase
|
162(2,2,2,2) |
Details |
|
catalase
|
134(1,2,4,14) |
Details |
|
guanylate cyclase
|
90(1,1,2,5) |
Details |
|
FKBP38
|
88(1,1,1,8) |
Details |
|
neurofibromin
|
86(1,1,1,6) |
Details |
|
interferon (protein family or complex)
|
86(1,1,1,6) |
Details |
|
phospholipase C
|
83(1,1,1,3) |
Details |
|
thrombin
|
81(1,1,1,1) |
Details |
|
protein its
|
81(1,1,1,1) |
Details |
|
cyclin
|
118(0,2,2,58) |
Details |
|
casein
|
108(0,2,8,18) |
Details |
|
cyclin D1
|
80(0,2,3,15) |
Details |
|
growth hormones
|
79(0,2,3,14) |
Details |
|
IKBKAP
|
93(0,1,10,18) |
Details |
|
per 1
|
69(0,1,7,9) |
Details |
|
bcl 2
|
48(0,1,2,13) |
Details |
|
ATPase
|
43(0,1,2,8) |
Details |
|
calcium channel (protein family or complex)
|
39(0,1,1,9) |
Details |
|
protein is
|
39(0,1,1,9) |
Details |
|
beta glucosidase
|
35(0,1,1,5) |
Details |
|
H1 histone (protein family or complex)
|
33(0,1,1,3) |
Details |
|
cAMP response element modulator
|
31(0,1,1,1) |
Details |
|
tip 2
|
31(0,1,1,1) |
Details |
|
cyclin dependent kinase (protein family or complex)
|
134(0,0,0,134) |
Details |
|
RIB1
|
44(0,0,5,19) |
Details |
|
CDK1
|
40(0,0,0,40) |
Details |
|
CDK2
|
34(0,0,0,34) |
Details |
|
cyclin A
|
26(0,0,0,26) |
Details |
|
delta aminolevulinic acid dehydratase
|
24(0,0,4,4) |
Details |
|
caspase 3
|
22(0,0,1,17) |
Details |
|
mitogen activated protein kinase (protein family or complex)
|
22(0,0,0,22) |
Details |
|
ACS1
|
19(0,0,1,14) |
Details |
|
alcohol dehydrogenase (protein family or complex)
|
19(0,0,3,4) |
Details |
|
CDK5
|
19(0,0,0,19) |
Details |
|
p21Cip1
|
18(0,0,1,13) |
Details |
|
E2F1
|
18(0,0,0,18) |
Details |
|
caspase (protein family or complex)
|
18(0,0,0,18) |
Details |
|
p34
|
16(0,0,0,16) |
Details |
|
beta glucuronidase
|
15(0,0,0,15) |
Details |
|
cytochrome c
|
15(0,0,0,15) |
Details |
|
P35
|
15(0,0,0,15) |
Details |
|
cyclin E
|
14(0,0,0,14) |
Details |
|
Adenine phosphoribosyltransferase
|
13(0,0,2,3) |
Details |
|
cyclin dependent protein kinases
|
12(0,0,0,12) |
Details |
|
P27
|
12(0,0,0,12) |
Details |
|
MAPK 2
|
11(0,0,0,11) |
Details |
|
p44mapk
|
11(0,0,0,11) |
Details |
|
p38
|
10(0,0,0,10) |
Details |
|
kinesin
|
9(0,0,0,9) |
Details |
|
involucrin
|
9(0,0,1,4) |
Details |
|
ClC 2
|
8(0,0,0,8) |
Details |
|
vimentin
|
8(0,0,0,8) |
Details |
|
malate dehydrogenase
|
8(0,0,1,3) |
Details |
|
mMDH
|
8(0,0,1,3) |
Details |
|
ABCG2
|
8(0,0,0,8) |
Details |
|
Sema3A
|
8(0,0,0,8) |
Details |
|
K14
|
8(0,0,1,3) |
Details |
|
Lipoxygenase (protein family or complex)
|
8(0,0,0,8) |
Details |
|
K10
|
8(0,0,1,3) |
Details |
|
pyrophosphatase
|
7(0,0,1,2) |
Details |
|
glycosyltransferases
|
7(0,0,1,2) |
Details |
|
fibrillin 1
|
7(0,0,1,2) |
Details |
|
Proton pumps
|
7(0,0,1,2) |
Details |
|
citron
|
7(0,0,1,2) |
Details |
|
cyclin B1
|
7(0,0,0,7) |
Details |
|
PP1
|
7(0,0,0,7) |
Details |
|
CYP1A1
|
7(0,0,0,7) |
Details |
|
filaggrin
|
6(0,0,1,1) |
Details |
|
protein phosphatases
|
6(0,0,0,6) |
Details |
|
Protein S
|
6(0,0,1,1) |
Details |
|
C18
|
6(0,0,1,1) |
Details |
|
elastin
|
6(0,0,1,1) |
Details |
|
ACO1
|
6(0,0,0,6) |
Details |
|
p82
|
6(0,0,0,6) |
Details |
|
CEPA
|
6(0,0,1,1) |
Details |
|
GLUT 1
|
6(0,0,1,1) |
Details |
|
cathepsin L
|
6(0,0,0,6) |
Details |
|
transforming growth factor beta 1
|
6(0,0,1,1) |
Details |
|
glycogen synthase kinase 3
|
6(0,0,0,6) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;