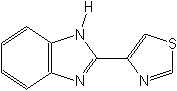
thiabendazole
Identification (Source:EPA,EU)
| ID |
500 |
| Name |
thiabendazole |
| CAS |
148-79-8 |
| IUPAC |
|
| Inchi |
InChI=1S/C10H7N3S/c1-2-4-8-7(3-1)12-10(13-8)9-5-14-6-11-9/h1-6H,(H,12,13) |
| CIPAC |
323 |
| EPA Code |
060101 |
| Formula |
C10H7N3S |
| SMILES |
n2c1c(cccc1)nc2c3ncsc3 |
| Status |
Approved |
Classification
| Mode of Action |
Systemic with curative and protective action. Acts by compromising the cytoskeleton through a selective interaction with 脽-tubulin |
| Pesticide Type |
Fungicide |
| Chemical Group |
Benzimidazole |
| Classification |
benzimidazole fungicides
thiazole fungicides
|
Properties (Source:PubChem)
| Molecular Weight |
201.25 |
| Physical State |
Colourless to white powder |
| AlogP |
2.154 |
| H-Bond Donor |
1 |
| H-Bond Acceptor |
2 |
| Surface Area |
184.06 |
| Polar Surface Area |
69.81 |
Structure
Ecotoxicology (Source:PPDB,IPM)
| Species |
Method |
Study Time |
Dose Type |
Value |
| Algae |
Chronic |
96 hour |
NOEC, growth |
3.2 mg l-1 |
| Algae |
Acute |
72 hour |
EC50, growth |
9.0 mg l-1 |
| Aquatic crustaceans |
Acute |
96 hour |
LC50 |
0.34 mg l-1 |
| Aquatic invertebrates |
Acute |
48 hour |
EC50 |
0.81 mg l-1 |
| Aquatic invertebrates |
Chronic |
21 day |
NOEC |
0.042 mg l-1 |
| Birds |
Acute |
|
LD50 |
> 2250 mg kg-1 |
| Birds |
Short term dietary |
|
LC50/LD50 |
> 5620 mg/kg feed |
| Earthworms |
Acute |
14 day |
LD50 |
> 1000 mg kg-1 |
| Earthworms |
Chronic |
14 day |
NOEC, reproduction |
4.2 mg kg-1 |
| Fish |
Acute |
96 hour |
LC50 |
0.55 mg l-1 |
| Fish |
Chronic |
21 day |
NOEC |
0.012 mg l-1 |
| Honeybees |
Acute |
48 hour |
LD50 |
50 μg bee-1 |
| Mammals |
Acute oral |
|
LD50 |
3100 mg kg-1 |
| Mammals |
Short term dietary |
|
NOEL |
> 10 mg kg-1 |
| Sediment dwelling organisms |
Chronic |
28 day |
NOEC, sediment |
2.0 mg kg-1 |
| Sediment dwelling organisms |
Chronic |
28 day |
NOEC, sediment |
3.0 mg kg-1 |
|
Bio-concentration factor |
|
CT50 |
Not available days |
|
Bio-concentration factor |
|
BCF |
96.5 |
Related targets
| Target Name |
RScore (About this table) |
|
|
CYP1A2
|
801(8,12,12,41) |
Details |
|
cytochrome P450 (protein family or complex)
|
330(3,5,7,20) |
Details |
|
cytochrome P450 1A1
|
282(2,4,9,37) |
Details |
|
carbonic anhydrase
|
195(2,3,3,5) |
Details |
|
aryl hydrocarbon receptor
|
148(1,3,3,8) |
Details |
|
superoxide dismutase
|
97(1,1,2,12) |
Details |
|
alpha tubulin
|
95(1,1,2,10) |
Details |
|
glutathione S transferase
|
85(1,1,1,5) |
Details |
|
methionine aminopeptidase
|
83(1,1,1,3) |
Details |
|
THBD
|
82(1,1,1,2) |
Details |
|
c fos
|
81(1,1,1,1) |
Details |
|
carbonic anhydrases (protein family or complex)
|
81(1,1,1,1) |
Details |
|
succinate dehydrogenase (protein family or complex)
|
71(0,2,3,6) |
Details |
|
IgE
|
83(0,1,4,38) |
Details |
|
gamma tubulin complex (protein family or complex)
|
55(0,1,2,20) |
Details |
|
cyclin
|
41(0,1,2,6) |
Details |
|
GF1
|
37(0,1,2,2) |
Details |
|
Acetylcholinesterase
|
37(0,1,1,7) |
Details |
|
malate dehydrogenase
|
36(0,1,1,6) |
Details |
|
TRF4
|
36(0,1,1,6) |
Details |
|
NPS1
|
35(0,1,1,5) |
Details |
|
Microsomal monooxygenase
|
33(0,1,1,3) |
Details |
|
CS2
|
33(0,1,1,3) |
Details |
|
CAPS
|
32(0,1,1,2) |
Details |
|
proton pump
|
32(0,1,1,2) |
Details |
|
GADD153
|
31(0,1,1,1) |
Details |
|
GRP78
|
31(0,1,1,1) |
Details |
|
acetylcholine receptors (protein family or complex)
|
31(0,1,1,1) |
Details |
|
thromboxane synthase
|
31(0,1,1,1) |
Details |
|
PF4
|
26(0,0,4,6) |
Details |
|
C18
|
23(0,0,2,13) |
Details |
|
arrestin
|
17(0,0,1,12) |
Details |
|
transducin
|
15(0,0,1,10) |
Details |
|
MSRB
|
15(0,0,2,5) |
Details |
|
P glycoproteins
|
14(0,0,0,14) |
Details |
|
interferon (protein family or complex)
|
13(0,0,2,3) |
Details |
|
CBF1
|
11(0,0,1,6) |
Details |
|
complex is
|
10(0,0,1,5) |
Details |
|
Dicer
|
10(0,0,1,5) |
Details |
|
interleukin 2
|
9(0,0,1,4) |
Details |
|
kinesin
|
9(0,0,0,9) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;