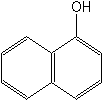
1-naphthol
Identification (Source:EPA,EU)
| ID |
1693 |
| Name |
1-naphthol |
| CAS |
90-15-3 |
| IUPAC |
1-naphthol |
| Inchi |
InChI=1S/C10H8O/c11-10-7-3-5-8-4-1-2-6-9(8)10/h1-7,11H |
| CIPAC |
|
| EPA Code |
|
| Formula |
C10H8O |
| SMILES |
O([H])c1c([H])c([H])c([H])c(c([H])c([H])c([H])c2[H])c12 |
| Status |
Not Available |
Classification
| Mode of Action |
|
| Pesticide Type |
|
| Chemical Group |
|
| Classification |
auxins
|
Properties (Source:PubChem)
| Molecular Weight |
144.16991 |
| Physical State |
|
| AlogP |
2.496 |
| H-Bond Donor |
1 |
| H-Bond Acceptor |
1 |
| Surface Area |
138.66 |
| Polar Surface Area |
20.23 |
Structure
Related targets
| Target Name |
RScore (About this table) |
|
|
cytochrome P450 (protein family or complex)
|
987(8,15,18,122) |
Details |
|
UGT1A6
|
765(6,12,16,85) |
Details |
|
UDPGT
|
586(4,10,13,71) |
Details |
|
immunoglobulins (protein family or complex)
|
463(4,8,9,18) |
Details |
|
protein a
|
408(3,8,8,18) |
Details |
|
Tyrosinase
|
337(3,6,6,7) |
Details |
|
transferase
|
456(2,9,15,56) |
Details |
|
phenol sulfotransferase
|
274(2,5,5,24) |
Details |
|
UGT1A1
|
251(2,4,5,26) |
Details |
|
alkaline phosphatase
|
232(2,3,5,32) |
Details |
|
SULT1A3
|
167(2,2,2,7) |
Details |
|
microsomal epoxide hydrolase
|
295(1,6,9,50) |
Details |
|
sulfotransferase
|
279(1,6,8,39) |
Details |
|
glutathione S transferase
|
266(1,5,8,51) |
Details |
|
Ugt1a9
|
206(1,4,5,31) |
Details |
|
GT1
|
159(1,3,4,14) |
Details |
|
menadione reductase
|
153(1,3,3,13) |
Details |
|
UGT1A4
|
141(1,2,4,21) |
Details |
|
udp glucuronosyltransferase 2B7
|
138(1,2,4,18) |
Details |
|
glial fibrillary acidic protein
|
126(1,2,3,11) |
Details |
|
anticholinesterase
|
121(1,2,2,11) |
Details |
|
hemoglobin (protein family or complex)
|
119(1,2,2,9) |
Details |
|
5 lipoxygenase
|
118(1,2,2,8) |
Details |
|
cytochrome c
|
118(1,2,2,8) |
Details |
|
cytochrome P450 reductase
|
118(1,2,2,8) |
Details |
|
CYP1A2
|
114(1,2,2,4) |
Details |
|
protein phosphatase inhibitor 1
|
112(1,2,2,2) |
Details |
|
xanthine oxidase
|
112(1,2,2,2) |
Details |
|
UGT1A3
|
98(1,1,2,13) |
Details |
|
UGT1A8
|
95(1,1,2,10) |
Details |
|
interleukin 2
|
95(1,1,1,15) |
Details |
|
P11
|
88(1,1,2,3) |
Details |
|
alpha tubulin
|
85(1,1,1,5) |
Details |
|
quinone oxidoreductase
|
85(1,1,1,5) |
Details |
|
UGT3A1
|
85(1,1,1,5) |
Details |
|
LHRH
|
85(1,1,1,5) |
Details |
|
INAP
|
84(1,1,1,4) |
Details |
|
superoxide dismutase
|
83(1,1,1,3) |
Details |
|
haptoglobin
|
82(1,1,1,2) |
Details |
|
thyroid hormone receptor (protein family or complex)
|
82(1,1,1,2) |
Details |
|
beta glucosidase
|
82(1,1,1,2) |
Details |
|
UDP glucuronosyltransferase
|
88(0,2,3,23) |
Details |
|
CYP3A4
|
73(0,2,2,13) |
Details |
|
Lipoxygenase (protein family or complex)
|
66(0,2,2,6) |
Details |
|
UGT1
|
54(0,1,1,24) |
Details |
|
acid phosphatase
|
43(0,1,2,8) |
Details |
|
UGT2B
|
42(0,1,1,12) |
Details |
|
CYP2E1
|
38(0,1,1,8) |
Details |
|
apolipoprotein E
|
38(0,1,1,8) |
Details |
|
UDP glucuronosyl transferase (protein family or complex)
|
37(0,1,2,2) |
Details |
|
interleukin 1
|
36(0,1,1,6) |
Details |
|
PK1
|
36(0,1,1,6) |
Details |
|
beta adrenergic receptor (protein family or complex)
|
35(0,1,1,5) |
Details |
|
cytochrome b5
|
35(0,1,1,5) |
Details |
|
N acetyltransferase
|
34(0,1,1,4) |
Details |
|
soluble epoxide hydrolase
|
34(0,1,1,4) |
Details |
|
FVIII
|
33(0,1,1,3) |
Details |
|
SM NS
|
33(0,1,1,3) |
Details |
|
desmin
|
33(0,1,1,3) |
Details |
|
protein phosphatase
|
32(0,1,1,2) |
Details |
|
UGT2B11
|
32(0,1,1,2) |
Details |
|
pancreatic alpha amylase
|
32(0,1,1,2) |
Details |
|
complex is
|
32(0,1,1,2) |
Details |
|
L10
|
31(0,1,1,1) |
Details |
|
them 4
|
31(0,1,1,1) |
Details |
|
Ag3
|
31(0,1,1,1) |
Details |
|
yl 1
|
31(0,1,1,1) |
Details |
|
HL2
|
31(0,1,1,1) |
Details |
|
carbonic anhydrase C
|
31(0,1,1,1) |
Details |
|
cis 2
|
31(0,1,1,1) |
Details |
|
protein tip
|
31(0,1,1,1) |
Details |
|
vimentin
|
31(0,1,1,1) |
Details |
|
glucuronosyltransferase p
|
31(0,1,1,1) |
Details |
|
UGT1*7
|
24(0,0,1,19) |
Details |
Docunmentation of Network Visuallization
 Network initializing...Please wait
Network initializing...Please wait
Legend:

: Current entry node;

: Target node;

: Compound node;

 Network initializing...Please wait
Network initializing...Please wait  : Current entry node;
: Current entry node;
 : Target node;
: Target node;
 : Compound node;
: Compound node;